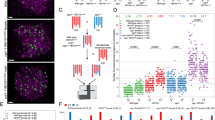
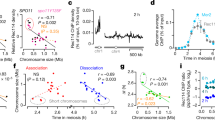
Abstract
Crossing-over ensures accurate chromosome segregation during meiosis, and every pair of chromosomes obtains at least one crossover, even though the majority of recombination sites yield non-crossovers. A putative regulator of crossing-over is RNF212, which is associated with variation in crossover rates in humans. We show that mouse RNF212 is essential for crossing-over, functioning to couple chromosome synapsis to the formation of crossover-specific recombination complexes. Selective localization of RNF212 to a subset of recombination sites is shown to be a key early step in the crossover designation process. RNF212 acts at these sites to stabilize meiosis-specific recombination factors, including the MutSγ complex (MSH4-MSH5). We infer that selective stabilization of key recombination proteins is a fundamental feature of meiotic crossover control. Haploinsufficiency indicates that RNF212 is a limiting factor for crossover control and raises the possibility that human alleles may alter the amount or stability of RNF212 and be risk factors for aneuploid conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hunter, N. Meiotic recombination. in Molecular Genetics of Recombination (eds. Aguilera, A. & Rothstein, R.) 381–442 (Springer-Verlag, Heidelberg, Germany, 2006).
Zickler, D. & Kleckner, N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999).
Sakuno, T., Tanaka, K., Hauf, S. & Watanabe, Y. Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev. Cell 21, 534–545 (2011).
Hirose, Y. et al. Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet. 7, e1001329 (2011).
Hassold, T., Hall, H. & Hunt, P. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 16 (Spec. No. 2), R203–R208 (2007).
Hassold, T. & Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001).
Keeney, S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2, 81–123 (2008).
Jones, G.H. The control of chiasma distribution. Symp. Soc. Exp. Biol. 38, 293–320 (1984).
Cole, F. et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430 (2012).
Allers, T. & Lichten, M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001).
Hunter, N. & Kleckner, N. The single-end invasion: an asymmetric intermediate at the double-strand break to double–Holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001).
Bishop, D.K. & Zickler, D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15 (2004).
Börner, G.V., Kleckner, N. & Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004).
Lynn, A., Soucek, R. & Borner, G.V. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15, 591–605 (2007).
Zakharyevich, K., Tang, S., Ma, Y. & Hunter, N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347 (2012).
Kong, A. et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319, 1398–1401 (2008).
Stefansson, H. et al. A common inversion under selection in Europeans. Nat. Genet. 37, 129–137 (2005).
Chowdhury, R., Bois, P.R., Feingold, E., Sherman, S.L. & Cheung, V.G. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 5, e1000648 (2009).
Fledel-Alon, A. et al. Variation in human recombination rates and its genetic determinants. PLoS ONE 6, e20321 (2011).
Fledel-Alon, A. et al. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 5, e1000658 (2009).
Kong, A. et al. Recombination rate and reproductive success in humans. Nat. Genet. 36, 1203–1206 (2004).
Ségurel, L., Leffler, E.M. & Przeworski, M. The case of the fickle fingers: how the PRDM9 zinc finger protein specifies meiotic recombination hotspots in humans. PLoS Biol. 9, e1001211 (2011).
Brick, K., Smagulova, F., Khil, P., Camerini-Otero, R.D. & Petukhova, G.V. Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012).
Grey, C. et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 9, e1001176 (2011).
Baudat, F. et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010).
Myers, S. et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879 (2010).
Hinch, A.G. et al. The landscape of recombination in African Americans. Nature 476, 170–175 (2011).
Agarwal, S. & Roeder, G.S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102, 245–255 (2000).
Jantsch, V. et al. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell Biol. 24, 7998–8006 (2004).
Deshaies, R.J. & Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Cheng, C.H. et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20, 2067–2081 (2006).
Perry, J., Kleckner, N. & Borner, G.V. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl. Acad. Sci. USA 102, 17594–17599 (2005).
Yuan, L. et al. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 5, 73–83 (2000).
Carlton, P.M. Three-dimensional structured illumination microscopy and its application to chromosome structure. Chromosome Res. 16, 351–365 (2008).
Moens, P.B., Marcon, E., Shore, J.S., Kochakpour, N. & Spyropoulos, B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 120, 1017–1027 (2007).
Snowden, T., Acharya, S., Butz, C., Berardini, M. & Fishel, R. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451 (2004).
Edelmann, W. et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21, 123–127 (1999).
Kneitz, B. et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097 (2000).
Kolas, N.K. & Cohen, P.E. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 107, 216–231 (2004).
Lipkin, S.M. et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31, 385–390 (2002).
Anderson, L.K., Reeves, A., Webb, L.M. & Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151, 1569–1579 (1999).
Baker, S.M. et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13, 336–342 (1996).
Edelmann, W. et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85, 1125–1134 (1996).
Baudat, F., Manova, K., Yuen, J.P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000).
Romanienko, P.J. & Camerini-Otero, R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 (2000).
de Vries, F.A. et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389 (2005).
Barchi, M. et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell Biol. 25, 7203–7215 (2005).
Di Giacomo, M. et al. Distinct DNA-damage–dependent and –independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl. Acad. Sci. USA 102, 737–742 (2005).
Kan, R. et al. Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol. Reprod. 78, 462–471 (2008).
Ward, J.O. et al. Mutation in mouse Hei10, an E3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3, e139 (2007).
Otto, S.P. et al. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 27, 358–367 (2011).
Cobb, J., Cargile, B. & Handel, M.A. Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev. Biol. 205, 49–64 (1999).
Holloway, J.K., Booth, J., Edelmann, W., McGowan, C.H. & Cohen, P.E. MUS81 generates a subset of MLH1-MLH3–independent crossovers in mammalian meiosis. PLoS Genet. 4, e1000186 (2008).
Marcon, E. & Moens, P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid–treated mouse spermatocytes. Genetics 165, 2283–2287 (2003).
Kolas, N.K. et al. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171, 447–458 (2005).
Ashley, T., Walpita, D. & de Rooij, D.G. Localization of two mammalian cyclin dependent kinases during mammalian meiosis. J. Cell Sci. 114, 685–693 (2001).
Woods, L.M. et al. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 145, 1395–1406 (1999).
Santucci-Darmanin, S. et al. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 14, 1539–1547 (2000).
Yang, F. et al. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 22, 682–691 (2008).
Adelman, C.A. & Petrini, J.H. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 4, e1000042 (2008).
Tsubouchi, T., Zhao, H. & Roeder, G.S. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev. Cell 10, 809–819 (2006).
Schwacha, A. & Kleckner, N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83, 783–791 (1995).
Oh, S.D. et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272 (2007).
Jessop, L., Rockmill, B., Roeder, G.S. & Lichten, M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2, e155 (2006).
Iyer, R.R., Pluciennik, A., Burdett, V. & Modrich, P.L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 106, 302–323 (2006).
Bhalla, N. & Dernburg, A.F. Prelude to a division. Annu. Rev. Cell Dev. Biol. 24, 397–424 (2008).
Bhalla, N., Wynne, D.J., Jantsch, V. & Dernburg, A.F. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 4, e1000235 (2008).
Macqueen, A.J. & Roeder, G.S. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr. Biol. 19, 1519–1526 (2009).
Yokoo, R. et al. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012).
Chelysheva, L. et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012).
Wang, K. et al. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8, e1002809 (2012).
Singh, M.K. et al. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene 26, 4825–4832 (2007).
Strong, E.R. & Schimenti, J.C. Evidence implicating CCNB1IP1, a RING domain–containing protein required for meiotic crossing over in mice, as an E3 SUMO ligase. Genes (Basel) 1, 440–451 (2010).
Nabeshima, K., Villeneuve, A.M. & Colaiacovo, M.P. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 168, 683–689 (2005).
Martinez-Perez, E. et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22, 2886–2901 (2008).
Kleckner, N. et al. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA 101, 12592–12597 (2004).
Zickler, D. From early homologue recognition to synaptonemal complex formation. Chromosoma 115, 158–174 (2006).
Henderson, K.A. & Keeney, S. Synaptonemal complex formation: where does it start? Bioessays 27, 995–998 (2005).
Anderson, L.K. & Stack, S.M. Recombination nodules in plants. Cytogenet. Genome Res. 109, 198–204 (2005).
Xu, K., Lu, T., Zhou, H., Bai, L. & Xiang, Y. The role of MSH5 C85T and MLH3 C2531T polymorphisms in the risk of male infertility with azoospermia or severe oligozoospermia. Clin. Chim. Acta 411, 49–52 (2010).
Terribas, E. et al. Changes in the expression profile of the meiosis-involved mismatch repair genes in impaired human spermatogenesis. J. Androl. 31, 346–357 (2010).
Hanks, M., Wurst, W., Anson-Cartwright, L., Auerbach, A.B. & Joyner, A.L. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science 269, 679–682 (1995).
Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J.C. Derivation of completely cell culture–derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90, 8424–8428 (1993).
Markel, P. et al. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 17, 280–284 (1997).
Susiarjo, M., Rubio, C. & Hunt, P. Analyzing mammalian female meiosis. Methods Mol. Biol. 558, 339–354 (2009).
Qiao, H., Lohmiller, L. & Anderson, L. Cohesin proteins load sequentially during prophase I in tomato primary microsporocytes. Chromosome Res. 19, 193–207 (2011).
Meuwissen, R.L. et al. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 11, 5091–5100 (1992).
Holloway, J.K., Morelli, M.A., Borst, P.L. & Cohen, P.E. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 188, 779–789 (2010).
Acknowledgements
We thank C. Heyting (Wageningen University) for antibodies to SYCP1 and SYCP3, S. Keeney and M. Jasin (Memorial Sloan-Kettering Cancer Center) for Spo11 knockout mice, M. Paddy for help with SIM imaging, J. Trimmer for help with imaging tissue sections and G. Coop for discussions. Knockout mice were generated in collaboration with the University of California, Davis, Mouse Biology Program (MBP). B.d.M. is supported by grants from the CNRS, Association pour la Recherche sur le Cancer and Agence Nationale de la Recherche (ANR-09-BLAN-0269-01). This work was supported by US National Institutes of Health (NIH) grant R01GM084955 to N.H. N.H. is an Early Career Scientist of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
A.R., N.J., H.Q., Y.Y., K.B., P.E.C. and N.H. conceived and designed the experiments. A.R., N.J., H.Q., Y.Y., J.K.C., K.B., J.K.H., B.d.M., F.B. and N.H. performed the experiments. A.R., H.Q., Y.Y., J.K.C., K.B., B.d.M. and N.H. analyzed the data. J.W. and C.H. contributed reagents, materials and/or analysis tools. A.R., H.Q., Y.Y., K.B. and N.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Table 1 (PDF 5242 kb)
Rights and permissions
About this article
Cite this article
Reynolds, A., Qiao, H., Yang, Y. et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 45, 269–278 (2013). https://doi.org/10.1038/ng.2541
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2541
This article is cited by
-
The RING Domain of Rice HEI10 is Essential for Male, But Not Female Fertility
Rice (2024)
-
A novel recombination protein C12ORF40/REDIC1 is required for meiotic crossover formation
Cell Discovery (2023)
-
Crossover patterning in plants
Plant Reproduction (2023)
-
Recombination rates in pigs differ between breeds, sexes and individuals, and are associated with the RNF212, SYCP2, PRDM7, MEI1 and MSH4 loci
Genetics Selection Evolution (2022)
-
Genetic variation in recombination rate in the pig
Genetics Selection Evolution (2021)