Abstract
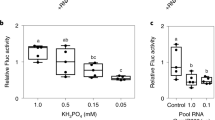
Effectors are essential virulence proteins produced by a broad range of parasites, including viruses, bacteria, fungi, oomycetes, protozoa, insects and nematodes. Upon entry into host cells, pathogen effectors manipulate specific physiological processes or signaling pathways to subvert host immunity. Most effectors, especially those of eukaryotic pathogens, remain functionally uncharacterized. Here, we show that two effectors from the oomycete plant pathogen Phytophthora sojae suppress RNA silencing in plants by inhibiting the biogenesis of small RNAs. Ectopic expression of these Phytophthora suppressors of RNA silencing enhances plant susceptibility to both a virus and Phytophthora, showing that some eukaryotic pathogens have evolved virulence proteins that target host RNA silencing processes to promote infection. These findings identify RNA silencing suppression as a common strategy used by pathogens across kingdoms to cause disease and are consistent with RNA silencing having key roles in host defense.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Haas, B.J. et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398 (2009).
Thines, M. & Kamoun, S. Oomycete-plant coevolution: recent advances and future prospects. Curr. Opin. Plant Biol. 13, 427–433 (2010).
Tyler, B.M. et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266 (2006).
Kale, S.D. et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142, 284–295 (2010).
Whisson, S.C. et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118 (2007).
Bozkurt, T.O., Schornack, S., Banfield, M.J. & Kamoun, S. Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492 (2012).
Chen, X. Small RNAs—secrets and surprises of the genome. Plant J. 61, 941–958 (2010).
Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 (2009).
Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644 (2010).
Vance, V. & Vaucheret, H. RNA silencing in plants—defense and counterdefense. Science 292, 2277–2280 (2001).
Katiyar-Agarwal, S. & Jin, H. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246 (2010).
Navarro, L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439 (2006).
Navarro, L., Jay, F., Nomura, K., He, S.Y. & Voinnet, O. Suppression of the microRNA pathway by bacterial effector proteins. Science 321, 964–967 (2008).
Ruiz, M.T., Voinnet, O. & Baulcombe, D.C. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946 (1998).
Brigneti, G. et al. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746 (1998).
Brosnan, C.A. & Voinnet, O. Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr. Opin. Plant Biol. 14, 580–587 (2011).
Dingwall, C. & Laskey, R.A. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16, 478–481 (1991).
Díaz-Pendon, J.A. & Ding, S.W. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326 (2008).
Chen, X., Liu, J., Cheng, Y. & Jia, D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129, 1085–1094 (2002).
Prigge, M.J. & Wagner, D.R. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1279 (2001).
Ren, G. et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. USA 109, 12817–12821 (2012).
Yu, B. et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 105, 10073–10078 (2008).
Li, F. et al. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 109, 1790–1795 (2012).
Zhai, J. et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553 (2011).
Haldar, K., Kamoun, S., Hiller, N.L., Bhattacharje, S. & van Ooij, C. Common infection strategies of pathogenic eukaryotes. Nat. Rev. Microbiol. 4, 922–931 (2006).
Wroblewski, T., Tomczak, A. & Michelmore, R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3, 259–273 (2005).
Jiang, R.H., Tripathy, S., Govers, F. & Tyler, B.M. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. USA 105, 4874–4879 (2008).
Earley, K.W. et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 (2006).
Zhou, H. et al. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe 9, 177–186 (2011).
Lu, R. et al. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 101, 15742–15747 (2004).
Zhou, H., Morgan, R.L., Guttman, D.S. & Ma, W. Allelic variants of the Pseudomonas syringae type III effector HopZ1 are differentially recognized by plant resistance systems. Mol. Plant Microbe Interact. 22, 176–189 (2009).
Clough, S.J. & Bent, A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Kurihara, Y., Takashi, Y. & Watanabe, Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212 (2006).
Pall, G.S., Codony-Servat, C., Byrne, J., Ritchie, L. & Hamilton, A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 35, e60 (2007).
Park, W., Li, J., Song, R., Messing, J. & Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495 (2002).
Ji, L. et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 7, e1001358 (2011).
Jones, L. et al. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11, 2291–2301 (1999).
Heese, A. et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104, 12217–12222 (2007).
Dou, D. et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20, 1118–1133 (2008).
Judelson, H.S., Tyler, B.M. & Michelmore, R.W. Transformation of the oomycete pathogen, Phytophthora infestans. Mol. Plant Microbe Interact. 4, 602–607 (1991).
Acknowledgements
We thank B. Tyler (Oregon State University) for providing soybean seeds and ten effector clones. S. Kamoun (The Sainsbury Laboratory) and M. Coffey (University of California, Riverside) kindly provided pGR106 and P. sojae strain P6497, respectively. We are indebted to S.-W. Ding for sharing viral suppressor constructs and thoughtful input. This work was supported by funds from the University of California, Riverside, to W.M. and X.C., National Science Foundation (NSF) grant IOS-0847870 to W.M. and US Department of Agriculture–National Institute of Food and Agriculture (USDA-NIFA) grants 2010-04209 and 2008-00694 to X.C. and H.S.J., respectively. L.L. was supported by a fellowship from the China Scholarship Council.
Author information
Authors and Affiliations
Contributions
W.M. and X.C. developed the concept. W.M., X.C., Y.Q., Y.W. and H.S.J. designed the experiments. Y.Q., L.L., Q. Xiong, C.F., J.W., J.S., X.W., X.L., Q. Xiang, S.J. and F.Z. performed the experiments. W.M., X.C. and Y.W. analyzed the data. W.M., X.C., H.S.J., Y.Q., L.L. and Y.W. wrote the manuscript. W.M. conceived, directed and coordinated the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Tables 1–3 (PDF 9930 kb)
Rights and permissions
About this article
Cite this article
Qiao, Y., Liu, L., Xiong, Q. et al. Oomycete pathogens encode RNA silencing suppressors. Nat Genet 45, 330–333 (2013). https://doi.org/10.1038/ng.2525
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2525
This article is cited by
-
Effector modularity promotes functional diversification and evolutionary processes
Science China Life Sciences (2023)
-
Designing Climate-Resilient Crops for Sustainable Agriculture: A Silent Approach
Journal of Plant Growth Regulation (2023)
-
Plant and animal small RNA communications between cells and organisms
Nature Reviews Molecular Cell Biology (2022)
-
Continuous monitoring of chemical signals in plants under stress
Nature Reviews Chemistry (2022)
-
Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems
Nature Reviews Molecular Cell Biology (2022)