Abstract
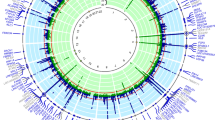
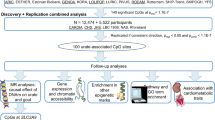
Elevated serum urate concentrations can cause gout, a prevalent and painful inflammatory arthritis. By combining data from >140,000 individuals of European ancestry within the Global Urate Genetics Consortium (GUGC), we identified and replicated 28 genome-wide significant loci in association with serum urate concentrations (18 new regions in or near TRIM46, INHBB, SFMBT1, TMEM171, VEGFA, BAZ1B, PRKAG2, STC1, HNF4G, A1CF, ATXN2, UBE2Q2, IGF1R, NFAT5, MAF, HLF, ACVR1B-ACVRL1 and B3GNT4). Associations for many of the loci were of similar magnitude in individuals of non-European ancestry. We further characterized these loci for associations with gout, transcript expression and the fractional excretion of urate. Network analyses implicate the inhibins-activins signaling pathways and glucose metabolism in systemic urate control. New candidate genes for serum urate concentration highlight the importance of metabolic control of urate production and excretion, which may have implications for the treatment and prevention of gout.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
So, A. & Thorens, B. Uric acid transport and disease. J. Clin. Invest. 120, 1791–1799 (2010).
Zhu, Y., Pandya, B.J. & Choi, H.K. Prevalence of gout and hyperuricemia in the US general population: the National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum. 63, 3136–3141 (2011).
Arromdee, E., Michet, C.J., Crowson, C.S., O′Fallon, W.M. & Gabriel, S.E. Epidemiology of gout: is the incidence rising? J. Rheumatol. 29, 2403–2406 (2002).
Wallace, K.L., Riedel, A.A., Joseph-Ridge, N. & Wortmann, R. Increasing prevalence of gout and hyperuricemia over 10 years among older adults in a managed care population. J. Rheumatol. 31, 1582–1587 (2004).
Yoo, H.G. et al. Prevalence of insulin resistance and metabolic syndrome in patients with gouty arthritis. Rheumatol. Int. 31, 485–491 (2011).
Singh, J.A. Quality of life and quality of care for patients with gout. Curr. Rheumatol. Rep. 11, 154–160 (2009).
Becker, M.A. et al. Quality of life and disability in patients with treatment-failure gout. J. Rheumatol. 36, 1041–1048 (2009).
Brook, R.A. et al. The economic burden of gout on an employed population. Curr. Med. Res. Opin. 22, 1381–1389 (2006).
Hediger, M.A., Johnson, R.J., Miyazaki, H. & Endou, H. Molecular physiology of urate transport. Physiology (Bethesda) 20, 125–133 (2005).
Whitfield, J.B. & Martin, N.G. Inheritance and alcohol as factors influencing plasma uric acid levels. Acta Genet. Med. Gemellol. (Roma) 32, 117–126 (1983).
Yang, Q. et al. Genome-wide search for genes affecting serum uric acid levels: the Framingham Heart Study. Metabolism 54, 1435–1441 (2005).
Nath, S.D. et al. Genome scan for determinants of serum uric acid variability. J. Am. Soc. Nephrol. 18, 3156–3163 (2007).
Li, S. et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 3, e194 (2007).
Doring, A. et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 40, 430–436 (2008).
Vitart, V. et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat. Genet. 40, 437–442 (2008).
Dehghan, A. et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet 372, 1953–1961 (2008).
Kolz, M. et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 5, e1000504 (2009).
Yang, Q. et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ. Cardiovasc. Genet. 3, 523–530 (2010).
Woodward, O.M. et al. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc. Natl. Acad. Sci. USA 106, 10338–10342 (2009).
Visscher, P.M., Brown, M.A., McCarthy, M.I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Suhre, K. et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 (2011).
Sato, M. et al. Renal secretion of uric acid by organic anion transporter 2 (OAT2/SLC22A7) in human. Biol. Pharm. Bull. 33, 498–503 (2010).
Wallace, S.L. et al. Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum. 20, 895–900 (1977).
Choi, H.K., Atkinson, K., Karlson, E.W., Willett, W. & Curhan, G. Purine-rich foods, dairy and protein intake, and the risk of gout in men. N. Engl. J. Med. 350, 1093–1103 (2004).
Ehret, G.B. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011).
Raychaudhuri, S. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009).
Ishizuka, T. et al. The human phosphoribosylpyrophosphate synthetase–associated protein 39 gene (PRPSAP1) is located in the chromosome region 17q24-q25. Genomics 33, 332–334 (1996).
Luo, Z., Saha, A.K., Xiang, X. & Ruderman, N.B. AMPK, the metabolic syndrome and cancer. Trends Pharmacol. Sci. 26, 69–76 (2005).
Tong, X., Zhao, F., Mancuso, A., Gruber, J.J. & Thompson, C.B. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc. Natl. Acad. Sci. USA 106, 21660–21665 (2009).
Levine, A.J. & Puzio-Kuter, A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330, 1340–1344 (2010).
Iynedjian, P.B. Molecular physiology of mammalian glucokinase. Cell. Mol. Life Sci. 66, 27–42 (2009).
Quinones Galvan, A. et al. Effect of insulin on uric acid excretion in humans. Am. J. Physiol. 268, E1–E5 (1995).
Ter Maaten, J.C. et al. Renal handling of urate and sodium during acute physiological hyperinsulinaemia in healthy subjects. Clin. Sci. (Lond.) 92, 51–58 (1997).
Becker, M.A. et al. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N. Engl. J. Med. 353, 2450–2461 (2005).
Iwatani, M. et al. Troglitazone decreases serum uric acid concentrations in type II diabetic patients and non-diabetics. Diabetologia 43, 814–815 (2000).
Gokcel, A. et al. Evaluation of the safety and efficacy of sibutramine, orlistat and metformin in the treatment of obesity. Diabetes Obes. Metab. 4, 49–55 (2002).
Seber, S., Ucak, S., Basat, O. & Altuntas, Y. The effect of dual PPARα/γ stimulation with combination of rosiglitazone and fenofibrate on metabolic parameters in type 2 diabetic patients. Diabetes Res. Clin. Pract. 71, 52–58 (2006).
Tsouli, S.G., Liberopoulos, E.N., Mikhailidis, D.P., Athyros, V.G. & Elisaf, M.S. Elevated serum uric acid levels in metabolic syndrome: an active component or an innocent bystander? Metabolism 55, 1293–1301 (2006).
Rizos, C.V., Liberopoulos, E.N., Mikhailidis, D.P. & Elisaf, M.S. Pleiotropic effects of thiazolidinediones. Expert Opin. Pharmacother. 9, 1087–1108 (2008).
Sulem, P. et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat. Genet. 43, 1127–1130 (2011).
The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am. J. Epidemiol. 129, 687–702 (1989).
Park, J.H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Yang, J., Lee, S.H., Goddard, M.E. & Visscher, P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Fuchsberger, C., Taliun, D., Pramstaller, P.P. & Pattaro, C. GWAtoolbox: an R package for fast quality control and handling of genome-wide association studies meta-analysis data. Bioinformatics 28, 444–445 (2012).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Pe'er, I., Yelensky, R., Altshuler, D. & Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Acknowledgements
A detailed list of acknowledgments is provided in the Supplementary Note.
Author information
Authors and Affiliations
Consortia
Contributions
Study design: A. Köttgen, C.G., E.A. and M. Caulfield.
Design and/or management of the individual studies: A.A.H., A. Tenesa, A.F.W., A.L., B.M.P., C.G., D.I.C., D.R., E.G.H., E.O., E. Trabetti, G.C., G.P., H. Campbell, H.-E.W., H. Snieder, I.J.D., J.A., J.C., J.F.W., J.V., L.M.R., M. Ciullo, M. Caulfield, M.F., M. Kubo, M.L., M.V., N.H., N.J.S., N. Kamatani, O.M.W., O.P., O.R., P.B.M., P.D., P.G., P.K., P. Mudgal, P.M.R., P.P.P., P.V., R.M.P., R. Sorice, S.H.W., S.M.F., S.U., T.E., T.L., Toshihiro Tanaka, V.S., W.H.L.K., Y.N., Y.O. and Z.K.
Phenotype collection: A.A.H., A.J.G., A.v.E., B.M.P., E.O., G.C., G.G., G.K.G., G.W., H. Snieder, I.J.D., I.K., I. Persico, J.C., J.F.W., J.V., L. Frogheri, M. Ciullo, M. Caulfield, M.G.D., M.G.P., M.J.B., M. Kähönen, M. Kubo, M.L., M. Pirastu, M.V., N.J.S., O.D., O.M.W., O.P., P. Sharma, P.B.M., P.K., P. Mudgal, P.P.P., P.V., S. Schipf, S.H.W., S.M.F., S.T., S.U., T. Zemunik and V.S.
Genotyping: A.A.H., A. Tenesa, A. Teumer, C. Hayward, D.I.C., E.L., F.E., G.C., G.D., G.W.M., I. Persico, J.F.W., M. Ciullo, M. Caulfield, M.E.K., M.G.D., M.G.P., M. Kubo, M.L., M. Putku, M.W., N.J.S., N. Klopp, O.R., P.B.M., P.D., P.M.R., P.P.P., P.v.d.H., R.J.S., S.M.F., T.E., T.L., T. Zeller and T. Zemunik.
Statistical methods and analysis: A. Tenesa, A. Tin, A. Köttgen, A. Teumer, A. Demirkan, C. Hayward, C. Hundertmark, C.G., C. Schurmann, D.C., D.I.C., D.R., E.A., E.G.H., F.M., F.T., G.A.T., G.K.G., G.L., G.M., G.P., I.M.L., I. Prokopenko, J.H., J.K., L.M.L., L.M.R., L.P., M.A.N., M. Steri, M. Bochud, M.E.K., M.F., M. Kähönen, M. Stumvoll, M. Putku, N.P., O.D., P. Mudgal, P.N., P.v.d.H., R.M.P., R.P.S.M., S.C., S.H.W., S. Sanna, T.E., T.H., T.L., V.V., W.H.L.K., X. Li, Y.O. and Z.K.
Interpretation of results: A. Tenesa, A. Tin, A. Köttgen, A.L.G., A. Teumer, B.M.P., C.G., D.R., E.A., G.W.M., H. Campbell, H. Snieder, J.K., M. Ciullo, M.A.N., M. Bochud, M. Caulfield, O.M.W., P.v.d.H., R.M.P., S.H.W., T.H., T.L., Toshihiro Tanaka, V.V., W.H.L.K., Y.O. and Z.K.
Manuscript review: A.A.H., A.B.S., A. Tenesa, A. Dehghan, A.D.J., A. Tin, A. Grotevendt, A. Goel, A.G.U., A.H., A.I., A. Jula, A. Köttgen, A.L., A.L.G., A. Kraja, A.M., A. Döring, A. Tönjes, A.P., A.R.S., A.S., A. Johansson, A. Teumer, A.V.S., B.B., B.H.R.W., B.M.P., B.O.B., B.R.W., B.W.P., C. Hundertmark, C. Hengstenberg, C. Sala, C.L., C.M., C.M.v.D., C.O., C.M.O., C.P.N., C. Schurmann, C.S.F., D.I.C., D.R., D.R.J., D.S.S., D.T., E.B., E.G.H., E. Theodoratou, F.C., F.E., F.R., F.T., G.A.T., G.C., G.G., G.N., G.W.M., H. Campbell, H. Choi, H. Schmidt, H.L.H., H.O., H. Snieder, H.V., H.W., I.B.B., I.K., I.M.L., I.M.N., I. Prokopenko, I.R., J.A., J.B.W., J.C., J.C.C., J.C.M.W., J.E.M., J.F.M., J.F.P., J.F.W., J.H.S., J.H.Z., J.I.R., J.K., J.S., J.S.K., J.V., K.B., K.L., K.S., K.T.K., L.J.L., L. Ferrucci, L.Y., M. Bruinenberg, M.A.N., M. Bochud, M. Caulfield, M. Ciullo, M.F., M.F.F., M.G.D., M.I., M. Burnier, M. Stumvoll, M. Kähönen, M. Kirin, M.M., M.N., M. Perola, M. Struchalin, M. Schallert, M.W., N.B.-N., N.G.M., N.J.W., N. Kamatani, N.M.P.-H., N.S., O.D., O.P., O.R., P. Sharma, P.F., P.K., P. McArdle, P.P.P., P. Salo, P.S.W., P.V., P.v.d.H., Q.Y., Q.Z., R. Schmidt, R.J.F.L., R.J.S., R.N., R.P.S., R. Sorice, S.B., S.H.W., S.J.L.B., S.K., S.L., S.R., S. Sanna, S.-Y.S., T.B.H., T.D.S., T.H., T.L., Toshiko Tanaka, T. Zemunik, U.G., V.G., V.L., V.V., W.H.L.K., W.H.v.G., W.M., W.Z., X. Liu, Y.N., Y.O. and Z.K.
Analysis group: A. Köttgen, A. Teumer, C.G., C. Hundertmark, C.S.F., D.R., E.A., G.P., J.K., Q.Y., T.H., Toshiko Tanaka, V.V. and W.H.L.K.
Writing group: A. Köttgen, A. Teumer, C.G., C.M.O., C.S.F., E.A., J.K., M. Bochud, M. Caulfield, M. Ciullo and V.V.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of contributing members appears in the Supplementary Note.
A list of contributing members appears in the Supplementary Note.
A list of contributing members appears in the Supplementary Note.
A list of contributing members appears in the Supplementary Note.
A list of contributing members appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1–3, 5, 7–9, 11, 14, 16 and 18 and Supplementary Note (PDF 4438 kb)
Supplementary Table 4
All SNPs Associated with Serum Urate at p<5*10-8 (XLSX 247 kb)
Supplementary Table 6
Separate Results from Discovery, Replication and Combined Analyses for SNP-Urate Associations (XLSX 26 kb)
Supplementary Table 10
All SNPs Associated with Gout at p<1*10-6 (XLSX 53 kb)
Supplementary Table 12
Overall and Sex-Specific Association between SNPs and Fractional Excretion of Uric Acid (FEUA) (XLSX 23 kb)
Supplementary Table 13
Associations of Urate-Associated SNPs in African Americans and Individuals of Indian and Japanese Ancestry (XLSX 20 kb)
Supplementary Table 15
Association of Replicated Urate-Associated SNPs with Related Phenotypes (XLSX 22 kb)
Supplementary Table 17
Functional Network Associations Underlying Supplementary Figure 8 and Supplementary Figure 9 (XLSX 23 kb)
Rights and permissions
About this article
Cite this article
Köttgen, A., Albrecht, E., Teumer, A. et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 45, 145–154 (2013). https://doi.org/10.1038/ng.2500
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2500
This article is cited by
-
The causal relationship between COVID-19 and estimated glomerular filtration rate: a bidirectional Mendelian randomization study
BMC Nephrology (2024)
-
Genome-wide association study of serum magnesium in type 2 diabetes
Genes & Nutrition (2024)
-
Should Glucokinase be Given a Chance in Diabetes Therapeutics? A Clinical-Pharmacological Review of Dorzagliatin and Lessons Learned So Far
Clinical Drug Investigation (2024)
-
GWAS-identified hyperuricemia-associated IGF1R variant rs6598541 has a limited role in urate mediated inflammation in human mononuclear cells
Scientific Reports (2024)
-
GLUT9 as a potential drug target for chronic kidney disease: Drug target validation by a Mendelian randomization study
Journal of Human Genetics (2023)