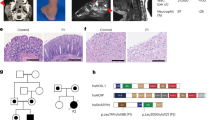
Abstract
Vici syndrome is a recessively inherited multisystem disorder characterized by callosal agenesis, cataracts, cardiomyopathy, combined immunodeficiency and hypopigmentation. To investigate the molecular basis of Vici syndrome, we carried out exome and Sanger sequence analysis in a cohort of 18 affected individuals. We identified recessive mutations in EPG5 (previously KIAA1632), indicating a causative role in Vici syndrome. EPG5 is the human homolog of the metazoan-specific autophagy gene epg-5, encoding a key autophagy regulator (ectopic P-granules autophagy protein 5) implicated in the formation of autolysosomes. Further studies showed a severe block in autophagosomal clearance in muscle and fibroblasts from individuals with mutant EPG5, resulting in the accumulation of autophagic cargo in autophagosomes. These findings position Vici syndrome as a paradigm of human multisystem disorders associated with defective autophagy and suggest a fundamental role of the autophagy pathway in the immune system and the anatomical and functional formation of organs such as the brain and heart.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Vici, C.D. et al. Agenesis of the corpus callosum, combined immunodeficiency, bilateral cataract, and hypopigmentation in two brothers. Am. J. Med. Genet. 29, 1–8 (1988).
del Campo, M. et al. Albinism and agenesis of the corpus callosum with profound developmental delay: Vici syndrome, evidence for autosomal recessive inheritance. Am. J. Med. Genet. 85, 479–485 (1999).
Chiyonobu, T. et al. Sister and brother with Vici syndrome: agenesis of the corpus callosum, albinism, and recurrent infections. Am. J. Med. Genet. 109, 61–66 (2002).
Miyata, R. et al. Sibling cases of Vici syndrome: sleep abnormalities and complications of renal tubular acidosis. Am. J. Med. Genet. A. 143, 189–194 (2007).
McClelland, V. et al. Vici syndrome associated with sensorineural hearing loss and evidence of neuromuscular involvement on muscle biopsy. Am. J. Med. Genet. A. 152A, 741–747 (2010).
Al-Owain, M. et al. Vici syndrome associated with unilateral lung hypoplasia and myopathy. Am. J. Med. Genet. A. 152A, 1849–1853 (2010).
Said, E., Soler, D. & Sewry, C. Vici syndrome—a rapidly progressive neurodegenerative disorder with hypopigmentation, immunodeficiency and myopathic changes on muscle biopsy. Am. J. Med. Genet. A. 158A, 440–444 (2012).
Finocchi, A. et al. Immunodeficiency in Vici syndrome: a heterogeneous phenotype. Am. J. Med. Genet. A. 158A, 434–439 (2012).
Rogers, C.R., Aufmuth, B. & Monesson, S. Vici Syndrome: a rare autosomal recessive syndrome with brain anomalies, cardiomyopathy, and severe intellectual disability. In Case Reports in Genetics Vol. 2011 1–4 (Hindawi Publishing Corporation, Cairo, NY, 2011).
Tian, Y. et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055 (2010).
Halama, N., Grauling-Halama, S.A., Beder, A. & Jager, D. Comparative integromics on the breast cancer–associated gene KIAA1632: clues to a cancer antigen domain. Int. J. Oncol. 31, 205–210 (2007).
Sjöblom, T. et al. The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 (2006).
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008).
Klionsky, D.J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937 (2007).
Mizushima, N. Autophagy: process and function. Genes Dev. 21, 2861–2873 (2007).
Maiuri, M.C., Zalckvar, E., Kimchi, A. & Kroemer, G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8, 741–752 (2007).
Rubinsztein, D.C., Gestwicki, J.E., Murphy, L.O. & Klionsky, D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 6, 304–312 (2007).
Klionsky, D.J. et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy 7, 1273–1294 (2010).
Mizushima, N. & Klionsky, D.J. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 27, 19–40 (2007).
Di Bartolomeo, S., Nazio, F. & Cecconi, F. The role of autophagy during development in higher eukaryotes. Traffic 11, 1280–1289 (2010).
Tra, T. et al. Autophagy in human embryonic stem cells. PLoS ONE 6, e27485 (2011).
Sandri, M. Autophagy in skeletal muscle. FEBS Lett. 584, 1411–1416 (2010).
Portbury, A.L., Willis, M.S. & Patterson, C. Tearin' up my heart: proteolysis in the cardiac sarcomere. J. Biol. Chem. 286, 9929–9934 (2011).
Cao, D.J., Gillette, T.G. & Hill, J.A. Cardiomyocyte autophagy: remodeling, repairing, and reconstructing the heart. Curr. Hypertens. Rep. 11, 406–411 (2009).
Li, W. et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J. Cell Biol. 197, 27–35 (2012).
Lange, S. et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science 308, 1599–1603 (2005).
Waters, S., Marchbank, K., Solomon, E., Whitehouse, C. & Gautel, M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 583, 1846–1852 (2009).
Kirkin, V., Lamark, T., Johansen, T. & Dikic, I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 5, 732–733 (2009).
Perera, S., Holt, M.R., Mankoo, B.S. & Gautel, M. Developmental regulation of MURF ubiquitin ligases and autophagy proteins nbr1, p62/SQSTM1 and LC3 during cardiac myofibril assembly and turnover. Dev. Biol. 351, 46–61 (2011).
Selcen, D. et al. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann. Neurol. 65, 83–89 (2009).
Edström, L., Thornell, L.E., Albo, J., Landin, S. & Samuelsson, M. Myopathy with respiratory failure and typical myofibrillar lesions. J. Neurol. Sci. 96, 211–228 (1990).
Masiero, E. & Sandri, M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 6, 307–309 (2010).
Taneike, M. et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 6, 600–606 (2010).
Klionsky, D.J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 (2012).
Schiaffino, S. & Mammucari, C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet. Muscle 1, 4 (2011).
Zhang, Y. et al. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 3, 337–346 (2007).
Nishino, I. et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406, 906–910 (2000).
Fukuda, T. et al. Autophagy and mistargeting of therapeutic enzyme in skeletal muscle in Pompe disease. Mol. Ther. 14, 831–839 (2006).
Lünemann, J.D. et al. β-amyloid is a substrate of autophagy in sporadic inclusion body myositis. Ann. Neurol. 61, 476–483 (2007).
Fujita, E. et al. Two endoplasmic reticulum–associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Hum. Mol. Genet. 16, 618–629 (2007).
Terman, A. & Brunk, U.T. Autophagy in cardiac myocyte homeostasis, aging, and pathology. Cardiovasc. Res. 68, 355–365 (2005).
Nakai, A. et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 13, 619–624 (2007).
Nishida, K., Kyoi, S., Yamaguchi, O., Sadoshima, J. & Otsu, K. The role of autophagy in the heart. Cell Death Differ. 16, 31–38 (2009).
Williams, A. et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr. Top. Dev. Biol. 76, 89–101 (2006).
Fimia, G.M. et al. Ambra1 regulates autophagy and development of the nervous system. Nature 447, 1121–1125 (2007).
Schmid, D. & Munz, C. Innate and adaptive immunity through autophagy. Immunity 27, 11–21 (2007).
Pua, H.H., Dzhagalov, I., Chuck, M., Mizushima, N. & He, Y.W. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204, 25–31 (2007).
Ganesan, A.K. et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 4, e1000298 (2008).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Li, K. & Stockwell, T.B. VariantClassifier: a hierarchical variant classifier for annotated genomes. BMC Res. Notes 3, 191 (2010).
Heath, K.E., Day, I.N. & Humphries, S.E. Universal primer quantitative fluorescent multiplex (UPQFM) PCR: a method to detect major and minor rearrangements of the low density lipoprotein receptor gene. J. Med. Genet. 37, 272–280 (2000).
Obermann, W.M. et al. The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J. Cell Biol. 134, 1441–1453 (1996).
Harlow, E. & Lane, D. Antibodies, a Laboratory Manual (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1988).
Acknowledgements
We are grateful to the individuals with Vici syndrome and their families for their participation in this study. We would like to thank our colleagues at the Genomics Facility of the Comprehensive Biomedical Research Centre of Guy's and St Thomas' NHS Foundation Trust for their support. We would also like to thank the physicians D. Creel, R.O. Hoffman and L. Al-Gazali for their input and productive discussions. H.J. was supported by a grant from the Guy's and St Thomas' Charitable Foundation (grant 070404). M.G. and A.L.K. were supported by the Leducq Foundation Transatlantic Network Proteotoxicity (11 CVD 04) and the Medical Research Council of Great Britain (MR/J010456/1). M.G. holds the British Heart Foundation Chair of Molecular Cardiology. H.J. would like to dedicate this work to the memory of Rahul Ghosh, his first patient with Vici syndrome.
Author information
Authors and Affiliations
Contributions
T.C. designed the experiments, performed whole-exome capture, Sanger sequencing, cDNA sequencing and quantitative PCR (qPCR) analysis, analyzed data and wrote the manuscript. A.L.K. and B.B. performed immunostaining, confocal microscopy, cell culture studies and protein blotting. Z.U. performed qPCR analysis. F.S., M.A.S., S.Y. and S.A. prepared and performed whole-exome capture and analyzed the exome sequencing data. C.D.-V., E.B., V.M., M.A.-O., S.K., C.K., G.F.H., F.A.W., A.E.t.H., R.C.R., D.M., R.M., M.H., E.S., D.S., P.M.K., C.W., F.M.F., S.A.-K., J.H. and M.D.C. provided clinical data. S.B., I.B., H.-H.G. and C.A.S. provided and analyzed neuropathological data. S.M. and D.J. provided clinical data and oversaw genetic aspects of the research. M.G. analyzed data obtained from immunostaining, confocal microscopy, cell culture studies and protein blotting and wrote the manuscript. H.J. provided clinical and neuropathological data, analyzed exome and Sanger sequencing data, oversaw all aspects of the research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–4 and Supplementary Figures 1–8 (PDF 1667 kb)
Rights and permissions
About this article
Cite this article
Cullup, T., Kho, A., Dionisi-Vici, C. et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet 45, 83–87 (2013). https://doi.org/10.1038/ng.2497
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2497
This article is cited by
-
Macroautophagy in CNS health and disease
Nature Reviews Neuroscience (2022)
-
A new UHPLC-MS/MS method for the screening of urinary oligosaccharides expands the detection of storage disorders
Orphanet Journal of Rare Diseases (2021)
-
Melatonin induces the rejuvenation of long-term ex vivo expanded periodontal ligament stem cells by modulating the autophagic process
Stem Cell Research & Therapy (2021)
-
Molecular and cellular basis of genetically inherited skeletal muscle disorders
Nature Reviews Molecular Cell Biology (2021)
-
Machinery, regulation and pathophysiological implications of autophagosome maturation
Nature Reviews Molecular Cell Biology (2021)