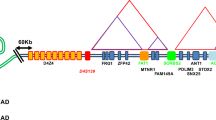
Abstract
Facioscapulohumeral dystrophy (FSHD) is characterized by chromatin relaxation of the D4Z4 macrosatellite array on chromosome 4 and expression of the D4Z4-encoded DUX4 gene in skeletal muscle. The more common form, autosomal dominant FSHD1, is caused by contraction of the D4Z4 array, whereas the genetic determinants and inheritance of D4Z4 array contraction–independent FSHD2 are unclear. Here, we show that mutations in SMCHD1 (encoding structural maintenance of chromosomes flexible hinge domain containing 1) on chromosome 18 reduce SMCHD1 protein levels and segregate with genome-wide D4Z4 CpG hypomethylation in human kindreds. FSHD2 occurs in individuals who inherited both the SMCHD1 mutation and a normal-sized D4Z4 array on a chromosome 4 haplotype permissive for DUX4 expression. Reducing SMCHD1 levels in skeletal muscle results in D4Z4 contraction–independent DUX4 expression. Our study identifies SMCHD1 as an epigenetic modifier of the D4Z4 metastable epiallele and as a causal genetic determinant of FSHD2 and possibly other human diseases subject to epigenetic regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Accessions
Ensembl
GenBank/EMBL/DDBJ
NCBI Reference Sequence
References
Statland, J.M. & Tawil, R. Facioscapulohumeral muscular dystrophy: molecular pathological advances and future directions. Curr. Opin. Neurol. 24, 423–428 (2011).
Balog, J. et al. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics 7, 579–584 (2012).
Bodega, B. et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 7, 41 (2009).
Cabianca, D.S. et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149, 819–831 (2012).
de Greef, J.C. et al. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 30, 1449–1459 (2009).
Jiang, G. et al. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 12, 2909–2921 (2003).
van Overveld, P.G. et al. Hypomethylation of D4Z4 in 4q-linked and non-4q–linked facioscapulohumeral muscular dystrophy. Nat. Genet. 35, 315–317 (2003).
Zeng, W. et al. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 5, e1000559 (2009).
Gabriëls, J. et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236, 25–32 (1999).
Hewitt, J.E. et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 3, 1287–1295 (1994).
Lyle, R., Wright, T.J., Clark, L.N. & Hewitt, J.E. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics 28, 389–397 (1995).
Snider, L. et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 18, 2414–2430 (2009).
Snider, L. et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6, e1001181 (2010).
van der Maarel, S.M., Tawil, R. & Tapscott, S.J. Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence. Trends Mol. Med. 17, 252–258 (2011).
Geng, L.N. et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51 (2012).
Bosnakovski, D. et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 27, 2766–2779 (2008).
Kowaljow, V. et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 17, 611–623 (2007).
Vanderplanck, C. et al. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS ONE 6, e26820 (2011).
Wallace, L.M. et al. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 69, 540–552 (2011).
Wuebbles, R.D., Long, S.W., Hanel, M.L. & Jones, P.L. Testing the effects of FSHD candidate gene expression in vertebrate muscle development. Int. J. Clin. Exp. Pathol. 3, 386–400 (2010).
van Deutekom, J.C. et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 2, 2037–2042 (1993).
Wijmenga, C. et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 2, 26–30 (1992).
Dixit, M. et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA 104, 18157–18162 (2007).
Lemmers, R.J. et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 32, 235–236 (2002).
Lemmers, R.J. et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653 (2010).
Spurlock, G., Jim, H.P. & Upadhyaya, M. Confirmation that the specific SSLP microsatellite allele 4qA161 segregates with fascioscapulohumeral muscular dystrophy (FSHD) in a cohort of multiplex and simplex FSHD families. Muscle Nerve 42, 820–821 (2010).
Thomas, N.S. et al. A large patient study confirming that facioscapulohumeral muscular dystrophy (FSHD) disease expression is almost exclusively associated with an FSHD locus located on a 4qA-defined 4qter subtelomere. J. Med. Genet. 44, 215–218 (2007).
Bamshad, M.J. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755 (2011).
Bhalla, N., Biggins, S. & Murray, A.W. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13, 632–645 (2002).
Lieb, J.D., Capowski, E.E., Meneely, P. & Meyer, B.J. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science 274, 1732–1736 (1996).
Chuang, P.T., Albertson, D.G. & Meyer, B.J. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79, 459–474 (1994).
Dej, K.J., Ahn, C. & Orr-Weaver, T.L. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics 168, 895–906 (2004).
Kanno, T. et al. A structural-maintenance-of-chromosomes hinge domain–containing protein is required for RNA-directed DNA methylation. Nat. Genet. 40, 670–675 (2008).
Blewitt, M.E. et al. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. USA 102, 7629–7634 (2005).
Blewitt, M.E. et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 40, 663–669 (2008).
Gendrel, A.V. et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell 23, 265–279 (2012).
Rakyan, V.K., Blewitt, M.E., Druker, R., Preis, J.I. & Whitelaw, E. Metastable epialleles in mammals. Trends Genet. 18, 348–351 (2002).
Duhl, D.M., Vrieling, H., Miller, K.A., Wolff, G.L. & Barsh, G.S. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8, 59–65 (1994).
van der Maarel, S.M., Frants, R.R. & Padberg, G.W. Facioscapulohumeral muscular dystrophy. Biochim. Biophys. Acta 1772, 186–194 (2007).
Scionti, I. et al. Facioscapulohumeral muscular dystrophy: new insights from compound heterozygotes and implication for prenatal genetic counselling. J. Med. Genet. 49, 171–178 (2012).
Lemmers, R.J. et al. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 81, 884–894 (2007).
Lemmers, R.J. et al. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 75, 1124–1130 (2004).
van Overveld, P.G. et al. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol. 58, 569–576 (2005).
de Greef, J.C. et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology 75, 1548–1554 (2010).
Lemmers, R.J.L. et al. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann. Neurol. 50, 816–819 (2001).
Lemmers, R.J.L.F. et al. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete duplication events in human evolution. Am. J. Hum. Genet. 86, 364–377 (2010).
Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Nelson, J.D., Denisenko, O. & Bomsztyk, K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 (2006).
Aartsma-Rus, A. Overview on AON design. Methods Mol. Biol. 867, 117–129 (2012).
Acknowledgements
The authors thank all subjects and family members for their participation. We thank D. Nickerson and J. Shendure for excellent assistance and B. Trask for helpful discussions and critical reading of the manuscript. This work was supported by grants from the US National Institutes of Health (NIH) (National Institute of Neurological Disorders and Stroke (NINDS) P01NS069539, Clinical & Translational Science Award (CTSA) UL1RR024160, National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) R01AR045203 and National Human Genome Research Institute (NHGRI) HG005608 and HG006493), a Netherlands Genomics Initiative (NGI) Horizon Valorization Project Grant (93515504), The University of Washington Center for Mendelian Genomics, the Muscular Dystrophy Association (MDA; 217596), the Fields Center for FSHD Research, the Geraldi Norton and Eklund family foundation, the FSH Society, The Friends of FSH Research, European Union Framework Programme 7 agreements 223026 (NMD-chip), 223143 (TechGene) and 2012-305121 (NEUROMICS) and the Stichting FSHD. Y.S. is supported by the China Scholarship Council.
Author information
Authors and Affiliations
Contributions
R.J.L.F.L., R.T., M.J.B., S.J.T., D.G.M., R.R.F., B.B., A.M.A.-R. and S.M.v.d.M. conceived of and designed the study. D.G.M., S.J.T. and S.M.v.d.M. directed the study. G.W.E.S., Y.S., Q.H. and D.G.M. performed the bioinformatics data analysis. R.T., B.G.M.v.E., G.W.P., S.S., C.D. and M.d.V. performed assessments of the FSHD2 phenotype. R.J.L.F.L., D.G.M., L.M.P., J.B., G.J.B., A.M.A., P.J.v.d.V., R.A., K.R.S., Y.D.K., R.K. and J.C.d.G. performed experiments. R.T., J.T.d.D., C.M.D.-S., G.W.P., B.G.M.v.E., G.N.F., M.d.V., C.D. and S.S. contributed samples, reagents, data and comments on the manuscript. R.J.L.F.L., S.J.T., D.G.M. and S.M.v.d.M. analyzed and interpreted data, and wrote the manuscript with the assistance and final approval of all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–3 and Supplementary Figures 1–4 (PDF 353 kb)
Rights and permissions
About this article
Cite this article
Lemmers, R., Tawil, R., Petek, L. et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 44, 1370–1374 (2012). https://doi.org/10.1038/ng.2454
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2454
This article is cited by
-
Flavones provide resistance to DUX4-induced toxicity via an mTor-independent mechanism
Cell Death & Disease (2023)
-
Management of spine deformity secondary to facioscapulohumeral dystrophy in pediatric patients. A case description and a literature review
Spine Deformity (2023)
-
Simultaneous measurement of the size and methylation of chromosome 4qA-D4Z4 repeats in facioscapulohumeral muscular dystrophy by long-read sequencing
Journal of Translational Medicine (2022)
-
Meeting report: the 2021 FSHD International Research Congress
Skeletal Muscle (2022)
-
Proximity ligation assay to detect DUX4 protein in FSHD1 muscle: a pilot study
BMC Research Notes (2022)