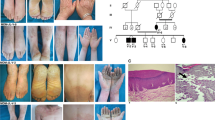
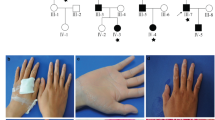
Abstract
Disseminated superficial actinic porokeratosis (DSAP) is an autosomal dominantly inherited epidermal keratinization disorder whose etiology remains unclear. We performed exome sequencing in one unaffected and two affected individuals from a DSAP family. The mevalonate kinase gene (MVK) emerged as the only candidate gene located in previously defined linkage regions after filtering against existing SNP databases, eight HapMap exomes and 1000 Genomes Project data and taking into consideration the functional implications of the mutations. Sanger sequencing in 57 individuals with familial DSAP and 25 individuals with sporadic DSAP identified MVK mutations in 33% and 16% of these individuals (cases), respectively. All 14 MVK mutations identified in our study were absent in 676 individuals without DSAP. Our functional studies in cultured primary keratinocytes suggest that MVK has a role in regulating calcium-induced keratinocyte differentiation and could protect keratinocytes from apoptosis induced by type A ultraviolet radiation. Our results should help advance the understanding of DSAP pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Chernosky, M.E. Porokeratosis. Arch. Dermatol. 122, 869–870 (1986).
Goerttler, E.A. & Jung, E.G. Porokeratosis Mibelli and skin carcinoma: a critical review. Humangenetik 26, 291–296 (1975).
Sasson, M. & Krain, A.D. Porokeratosis and cutaneous malignancy. Dermatol. Surg. 22, 339–342 (1996).
Otsuka, F., Someya, T. & Ishibashi, Y. Porokeratosis and malignant skin tumors. J. Cancer Res. Clin. Oncol. 117, 55–60 (1991).
Xia, J.H. et al. Identification of a locus for disseminated superficial actinic porokeratosis at chromosome 12q23.2–24.1. J. Invest. Dermatol. 114, 1071–1074 (2000).
Anderson, D.E. & Chernosky, M.E. Disseminated superficial actinic porokeratosis. Genetic aspects. Arch. Dermatol. 99, 408–412 (1969).
Zhang, Z.H. et al. A mutation in SART3 gene in a Chinese pedigree with disseminated superficial actinic porokeratosis. Br. J. Dermatol. 152, 658–663 (2005).
Xia, K. et al. A novel locus (DSAP2) for disseminated superficial actinic porokeratosis maps to chromosome 15q25.1–26.1. Br. J. Dermatol. 147, 650–654 (2002).
Liu, P. et al. Identification of a genetic locus for autosomal dominant disseminated superficial actinic porokeratosis on chromosome 1p31.3–p31.1. Hum. Genet. 123, 507–513 (2008).
Luan, J. et al. A novel locus for disseminated superficial actinic porokeratosis maps to chromosome 16q24.1–24.3. Hum. Genet. 129, 329–334 (2011).
Zhang, Z. et al. Fine mapping and identification of a candidate gene SSH1 in disseminated superficial actinic porokeratosis. Hum. Mutat. 24, 438 (2004).
Frank, J., van Geel, M. & van Steensel, M.A. Loss of heterozygosity studies on chromosome 12q in disseminated superficial actinic porokeratosis: lessons to be learned. J. Invest. Dermatol. 127, 2058–2059 (2007).
Frank, J., van Steensel, M.A. & van Geel, M. Lack of SSH1 mutations in Dutch patients with disseminated superficial actinic porokeratosis: is there really an association? Hum. Mutat. 28, 1241–1242 (2007).
Ng, S.B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Cooper, G.M. et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat. Methods 7, 250–251 (2010).
Wu, L.Q. et al. Confirmation and refinement of a genetic locus for disseminated superficial actinic porokeratosis (DSAP1) at 12q23.2–24.1. Br. J. Dermatol. 150, 999–1004 (2004).
Fu, Z., Wang, M., Potter, D., Miziorko, H.M. & Kim, J.J. The structure of a binary complex between a mammalian mevalonate kinase and ATP: insights into the reaction mechanism and human inherited disease. J. Biol. Chem. 277, 18134–18142 (2002).
Houten, S.M., Wanders, R.J. & Waterham, H.R. Biochemical and genetic aspects of mevalonate kinase and its deficiency. Biochim. Biophys. Acta 1529, 19–32 (2000).
McTaggart, S.J. Isoprenylated proteins. Cell. Mol. Life Sci. 63, 255–267 (2006).
Bouwstra, J.A. & Ponec, M. The skin barrier in healthy and diseased state. Biochim. Biophys. Acta 1758, 2080–2095 (2006).
Zhao, Y., Gartner, U., Smith, F.J. & McLean, W.H. Statins downregulate K6a promoter activity: a possible therapeutic avenue for pachyonychia congenita. J. Invest. Dermatol. 131, 1045–1052 (2011).
Edmondson, S.R., Werther, G.A. & Wraight, C.J. Calcium regulates the expression of insulin-like growth factor binding protein-3 by the human keratinocyte cell line HaCaT. J. Invest. Dermatol. 116, 491–497 (2001).
Shen, C.S. et al. Premature apoptosis of keratinocytes and the dysregulation of keratinization in porokeratosis. Br. J. Dermatol. 147, 498–502 (2002).
Haas, D. & Hoffmann, G.F. Mevalonate kinase deficiencies: from mevalonic aciduria to hyperimmunoglobulinemia D syndrome. Orphanet J. Rare Dis. 1, 13 (2006).
Cuisset, L. et al. Molecular analysis of MVK mutations and enzymatic activity in hyper-IgD and periodic fever syndrome. Eur. J. Hum. Genet. 9, 260–266 (2001).
Mandey, S.H., Schneiders, M.S., Koster, J. & Waterham, H.R. Mutational spectrum and genotype-phenotype correlations in mevalonate kinase deficiency. Hum. Mutat. 27, 796–802 (2006).
Drenth, J.P., Haagsma, C.J. & van der Meer, J.W. Hyperimmunoglobulinemia D and periodic fever syndrome. The clinical spectrum in a series of 50 patients. International Hyper-IgD Study Group. Medicine (Baltimore) 73, 133–144 (1994).
Cuisset, L. et al. Molecular analysis of MVK mutations and enzymatic activity in hyper-IgD and periodic fever syndrome. Eur. J. Hum. Genet. 9, 260–266 (2001).
Bader-Meunier, B. et al. Mevalonate kinase deficiency: a survey of 50 patients. Pediatrics 128, e152–e159 (2011).
Li, R., Li, Y., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Li, R. et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Acknowledgements
We thank the individuals and their families who participated in this project. This study was funded by Ministry of Education of China (IRT-1046).
Author information
Authors and Affiliations
Contributions
X.-J.Z., S.Y. and Jun Wang conceived of this study and obtained financial support. X.-J.Z., L.-D.S., Y.C., M.G. and X.F. participated in the design of the study and project management. S.-C.W., Z.-M.H., Z.-Y.W., Ping Li, B.-Y.W., W.W., X.-F.Y., J.-S.H., L.Z. and K.X. performed blood sample collection and selection. F.-S.Z., G.C., G.-Q.S., Y.X., J.-C.H. and T.-T.H. performed exome capture and paired-end sequencing. T.J., X.-B.Z., Y.-R.L., H.-M.Y. and Jian Wang performed next-generation sequencing analysis and filtering. X.Z., X.-F.T., W.-W.H., W.-J.W., Y.-J.Z., Jing Wang, F.-F.Q., B.L., J.Z., Z.-Y.Y., H.-F.Z., X.H., K.Y., Jie Wang and S.-X.Z. performed PCR and Sanger sequencing. X.-Y.Y., Y. Liu, B.-R.G., X.-D.Z., P.-G.W., H.L., S.-X.L., S.-K.S. and M.Z. performed other data analyses. S.-Q.Z., M.L., Y.-Q.R., H.C., Y. Li, H.-Y.T., Y.-T.D., J.-B.W., Y.-J.S., J.-P.G., Pan Li and F.-L.X. performed the cell culture and protein expression studies, gene sequencing, protein blot analysis and other function studies. M.L., X.Z., Y.-W.Z., J.-J.L. and W.-D.D. wrote and revised the manuscript. All the authors contributed to the final paper, with X.-J.Z., Jun Wang, S.-Q.Z., T.J., M.L., X.Z. and L.-D.S. having key roles.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–5 and Supplementary Note. (PDF 1266 kb)
Rights and permissions
About this article
Cite this article
Zhang, SQ., Jiang, T., Li, M. et al. Exome sequencing identifies MVK mutations in disseminated superficial actinic porokeratosis. Nat Genet 44, 1156–1160 (2012). https://doi.org/10.1038/ng.2409
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2409
This article is cited by
-
MicroRNA-874 targets phosphomevalonate kinase and inhibits cancer cell growth via the mevalonate pathway
Scientific Reports (2022)
-
Molecular mechanisms of phenotypic variability in monogenic autoinflammatory diseases
Nature Reviews Rheumatology (2021)
-
Update on the Genetics of Autoinflammatory Disorders
Current Allergy and Asthma Reports (2019)
-
The monogenic autoinflammatory diseases define new pathways in human innate immunity and inflammation
Nature Immunology (2017)