Abstract
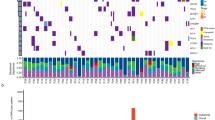
Small-cell lung cancer (SCLC) is an exceptionally aggressive disease with poor prognosis. Here, we obtained exome, transcriptome and copy-number alteration data from approximately 53 samples consisting of 36 primary human SCLC and normal tissue pairs and 17 matched SCLC and lymphoblastoid cell lines. We also obtained data for 4 primary tumors and 23 SCLC cell lines. We identified 22 significantly mutated genes in SCLC, including genes encoding kinases, G protein–coupled receptors and chromatin-modifying proteins. We found that several members of the SOX family of genes were mutated in SCLC. We also found SOX2 amplification in ∼27% of the samples. Suppression of SOX2 using shRNAs blocked proliferation of SOX2-amplified SCLC lines. RNA sequencing identified multiple fusion transcripts and a recurrent RLF-MYCL1 fusion. Silencing of MYCL1 in SCLC cell lines that had the RLF-MYCL1 fusion decreased cell proliferation. These data provide an in-depth view of the spectrum of genomic alterations in SCLC and identify several potential targets for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Siegel, R., Ward, E., Brawley, O. & Jemal, A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 61, 212–236 (2011).
Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics, 2012. CA Cancer J. Clin. 62, 10–29 (2012).
Hann, C.L. & Rudin, C.M. Fast, hungry and unstable: finding the Achilles' heel of small-cell lung cancer. Trends Mol. Med. 13, 150–157 (2007).
Wistuba, I.I., Gazdar, A.F. & Minna, J.D. Molecular genetics of small cell lung carcinoma. Semin. Oncol. 28, 3–13 (2001).
Mori, N. et al. Variable mutations of the RB gene in small-cell lung carcinoma. Oncogene 5, 1713–1717 (1990).
Arriola, E. et al. Genetic changes in small cell lung carcinoma. Clin. Transl. Oncol. 10, 189–197 (2008).
Yokomizo, A. et al. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene 17, 475–479 (1998).
Tatematsu, A. et al. Epidermal growth factor receptor mutations in small cell lung cancer. Clin. Cancer Res. 14, 6092–6096 (2008).
Shibata, T., Kokubu, A., Tsuta, K. & Hirohashi, S. Oncogenic mutation of PIK3CA in small cell lung carcinoma: a potential therapeutic target pathway for chemotherapy-resistant lung cancer. Cancer Lett. 283, 203–211 (2009).
Onuki, N. et al. Genetic changes in the spectrum of neuroendocrine lung tumors. Cancer 85, 600–607 (1999).
Sher, T., Dy, G.K. & Adjei, A.A. Small cell lung cancer. Mayo Clin. Proc. 83, 355–367 (2008).
Forbes, S.A. et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 38, D652–D657 (2010).
Pleasance, E.D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 (2010).
Ng, P.C. & Henikoff, S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 12, 436–446 (2002).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
González-Pérez, A. & Lopez-Bigas, N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 88, 440–449 (2011).
Kan, Z. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010).
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature published online, doi:10.1038/nature11282 (15 August 2012).
Molderings, G.J. et al. Multiple novel alterations in Kit tyrosine kinase in patients with gastrointestinally pronounced systemic mast cell activation disorder. Scand. J. Gastroenterol. 42, 1045–1053 (2007).
D'Angelo, S.P. & Pietanza, M.C. The molecular pathogenesis of small cell lung cancer. Cancer Biol. Ther. 10, 1–10 (2010).
Medina, P.P. et al. The SRY–HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum. Mol. Genet. 18, 1343–1352 (2009).
Tompkins, D.H. et al. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS ONE 4, e8248 (2009).
Wegner, M. SOX after SOX: SOXession regulates neurogenesis. Genes Dev. 25, 2423–2428 (2011).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Lujan, E., Chanda, S., Ahlenius, H., Sudhof, T.C. & Wernig, M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. USA 109, 2527–2532 (2012).
Bass, A.J. et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 41, 1238–1242 (2009).
Mäkelä, T.P., Saksela, K., Evan, G. & Alitalo, K. A fusion protein formed by L-myc and a novel gene in SCLC. EMBO J. 10, 1331–1335 (1991).
Robinson, D.R. et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 17, 1646–1651 (2011).
Lu, Y. et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE 5, e11022 (2010).
Gontan, C. et al. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev. Biol. 317, 296–309 (2008).
Sholl, L.M., Long, K.B. & Hornick, J.L. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl. Immunohistochem. Mol. Morphol. 18, 55–61 (2010).
Güre, A.O. et al. Serological identification of embryonic neural proteins as highly immunogenic tumor antigens in small cell lung cancer. Proc. Natl. Acad. Sci. USA 97, 4198–4203 (2000).
Kwak, E.L. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703 (2010).
Bergethon, K. et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 30, 863–870 (2012).
Takeuchi, K. et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 18, 378–381 (2012).
Morgan, M. et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25, 2607–2608 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Martincorena, I., Seshasayee, A.S. & Luscombe, N.M. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature 485, 95–98 (2012).
Barbieri, C.E. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012).
Greenman, C.D. et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 11, 164–175 (2010).
Mermel, C.H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Simms, E., Gazdar, A.F., Abrams, P.G. & Minna, J.D. Growth of human small cell (oat cell) carcinoma of the lung in serum-free growth factor–supplemented medium. Cancer Res. 40, 4356–4363 (1980).
Phelps, R.M. et al. NCI–Navy Medical Oncology Branch cell line data base. J. Cell. Biochem. Suppl. 24, 32–91 (1996).
Carney, D.N., Bepler, G. & Gazdar, A.F. The serum-free establishment and in vitro growth properties of classic and variant small cell lung cancer cell lines. Recent Results Cancer Res. 99, 157–166 (1985).
Sarbassov, D.D., Guertin, D.A., Ali, S.M. & Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005).
Wiederschain, D. et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8, 498–504 (2009).
Acknowledgements
The authors would like to thank Genentech DNA Sequencing and Oligo groups for their help with the project. We thank M.A. Huntley and J. Degenhardt for bioinformatics support and the Pathology Core Labs for providing histology, IHC and tissue management support. This work was also supported by grants from the Burroughs Wellcome Fund, the Flight Attendant Medical Research Institute, the Johns Hopkins Specialized Programs of Research Excellence (SPORE) NCI P50CA058184 (M.V.B. and C.M.R.), the Colorado SPORE NCI P50 CA058187 (M.V.-G) and the University of Texas SPORE NCI P50CA70907 (J.D.M., A.F.G. and K.E.H.). D.D.P. is supported by the Coordenacao de Aperfeicoamento de Passoal de Nivel Superior (CAPES) Foundation and the Ministry of Education of Brazil.
Author information
Authors and Affiliations
Contributions
C.M.R. and S.S. conceived the study and designed the experiments. E.W.S. and S.D. performed the exome and whole-genome sequencing, RNA-seq and copy-number analysis. Z.M. and Y.G. performed validation of the fusions. Z.M. managed exome capture. J.T.P., E.A.B., S.C., V.J., B.S.J., W.Y. and C.P. performed biological validated studies. J. Shin, D.D.P., P.B.I. and M.V.-G. performed SOX2 IHC and FISH studies. K.E.H., A.F.G. and J.D.M. provided reagents and analysis support. J. Stinson, C.K.F., D.B., C.S.R. and J.G. collected sequencing data and performed mutation validation. F.G. and Z.Z. predicted the functional effects of mutations. E.W.S., P.M.H., R.B., T.D.W. and R.G. provided bioinformatics support, including the algorithm for variant calling, fusion detection and copy-number calling. R.B. and P.M.H. analyzed SNP array data. H.K., H.M.S., P.B.I., M.V.B. and A.F.G. provided pathology support. F.J.d.S., D.S.S., R.L.Y. and J.D.M. provided critical analysis and organizational support. D.S.S., E.W.S., S.D., Z.M., C.M.R. and J.T.P. wrote the manuscript, which was reviewed and edited by the other coauthors.
Corresponding authors
Ethics declarations
Competing interests
C.M.R. has previously consulted for Genentech. P.B.I. is a consultant for Leica Microsystems, the manufacturer of a device used in this study. The terms of these arrangements are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. Some of the authors, as indicated in the author affiliations, are employees of Genentech and hold shares in Roche.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 816 kb)
Supplementary Tables
Supplementary Tables 1–13 (XLS 5612 kb)
Rights and permissions
About this article
Cite this article
Rudin, C., Durinck, S., Stawiski, E. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44, 1111–1116 (2012). https://doi.org/10.1038/ng.2405
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2405
This article is cited by
-
Genetically-engineered mouse models of small cell lung cancer: the next generation
Oncogene (2024)
-
Inhibiting NR5A2 targets stemness in pancreatic cancer by disrupting SOX2/MYC signaling and restoring chemosensitivity
Journal of Experimental & Clinical Cancer Research (2023)
-
MYC drives platinum resistant SCLC that is overcome by the dual PI3K-HDAC inhibitor fimepinostat
Journal of Experimental & Clinical Cancer Research (2023)
-
Expression of down-regulated ERV LTR elements associates with immune activation in human small-cell lung cancers
Mobile DNA (2023)
-
Molecular features and evolutionary trajectory of ASCL1+ and NEUROD1+ SCLC cells
British Journal of Cancer (2023)