Abstract
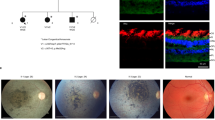
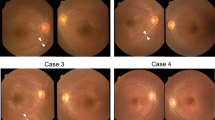
Leber congenital amaurosis (LCA) is an infantile-onset form of inherited retinal degeneration characterized by severe vision loss1,2. Two-thirds of LCA cases are caused by mutations in 17 known disease-associated genes3 (Retinal Information Network (RetNet)). Using exome sequencing we identified a homozygous missense mutation (c.25G>A, p.Val9Met) in NMNAT1 that is likely to be disease causing in two siblings of a consanguineous Pakistani kindred affected by LCA. This mutation segregated with disease in the kindred, including in three other children with LCA. NMNAT1 resides in the previously identified LCA9 locus and encodes the nuclear isoform of nicotinamide mononucleotide adenylyltransferase, a rate-limiting enzyme in nicotinamide adenine dinucleotide (NAD+) biosynthesis4,5. Functional studies showed that the p.Val9Met alteration decreased NMNAT1 enzyme activity. Sequencing NMNAT1 in 284 unrelated families with LCA identified 14 rare mutations in 13 additional affected individuals. These results are the first to link an NMNAT isoform to disease in humans and indicate that NMNAT1 mutations cause LCA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Weleber, R.G. Infantile and childhood retinal blindness: a molecular perspective. Ophthalmic Genet. 23, 71–97 (2002).
Michaelides, M., Hardcastle, A.J., Hunt, D.M. & Moore, A.T. Progressive cone and cone-rod dystrophies: phenotypes and underlying molecular genetic basis. Surv. Ophthalmol. 51, 232–258 (2006).
den Hollander, A.I., Black, A., Bennett, J. & Cremers, F.P. Lighting a candle in the dark: advances in genetics and gene therapy of recessive retinal dystrophies. J. Clin. Invest. 120, 3042–3053 (2010).
Keen, T.J. et al. Identification of a locus (LCA9) for Leber's congenital amaurosis on chromosome 1p36. Eur. J. Hum. Genet. 11, 420–423 (2003).
Lau, C., Niere, M. & Ziegler, M. The NMN/NaMN adenylyltransferase (NMNAT) protein family. Front. Biosci. 14, 410–431 (2009).
Pierce, E.A. Pathways to photoreceptor cell death in inherited retinal degenerations. Bioessays 23, 605–618 (2001).
Daiger, S.P., Bowne, S.J. & Sullivan, L.S. Perspective on genes and mutations causing retinitis pigmentosa. Arch. Ophthalmol. 125, 151–158 (2007).
Maguire, A.M. et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 358, 2240–2248 (2008).
Bainbridge, J.W. et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med. 358, 2231–2239 (2008).
Cideciyan, A.V. et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc. Natl. Acad. Sci. USA 105, 15112–15117 (2008).
Maguire, A.M. et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet 374, 1597–1605 (2009).
Jacobson, S.G. et al. Gene therapy for Leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. Arch. Ophthalmol. 130, 9–24 (2012).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Grant, G.R. et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 27, 2518–2528 (2011).
Ramensky, V., Bork, P. & Sunyaev, S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900 (2002).
Ng, P.C. & Henikoff, S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Sullivan, L.S. et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest. Ophthalmol. Vis. Sci. 47, 3052–3064 (2006).
Stone, E.M. Leber congenital amaurosis—a model for efficient genetic testing of heterogeneous disorders: LXIV Edward Jackson Memorial Lecture. Am. J. Ophthalmol. 144, 791–811 (2007).
Wang, M. & Marin, A. Characterization and prediction of alternative splice sites. Gene 366, 219–227 (2006).
Belenky, P., Bogan, K.L. & Brenner, C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 32, 12–19 (2007).
Berger, F., Lau, C., Dahlmann, M. & Ziegler, M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 280, 36334–36341 (2005).
Coleman, M.P. & Freeman, M.R. Wallerian degeneration, wlds, and nmnat. Annu. Rev. Neurosci. 33, 245–267 (2010).
Conforti, L. et al. Reducing expression of NAD+ synthesizing enzyme NMNAT1 does not affect the rate of Wallerian degeneration. FEBS J. 278, 2666–2679 (2011).
Zhai, R.G. et al. Drosophila NMNAT maintains neural integrity independent of its NAD synthesis activity. PLoS Biol. 4, e416 (2006).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Worth, C.L., Preissner, R. & Blundell, T.L. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 39, W215–W222 (2011).
Bogan, K.L. & Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130 (2008).
Nikiforov, A., Dolle, C., Niere, M. & Ziegler, M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 286, 21767–21778 (2011).
Conforti, L. et al. WldS protein requires Nmnat activity and a short N-terminal sequence to protect axons in mice. J. Cell Biol. 184, 491–500 (2009).
Avery, M.A., Sheehan, A.E., Kerr, K.S., Wang, J. & Freeman, M.R. WldS requires Nmnat1 enzymatic activity and N16-VCP interactions to suppress Wallerian degeneration. J. Cell Biol. 184, 501–513 (2009).
Berger, F., Lau, C. & Ziegler, M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc. Natl. Acad. Sci. USA 104, 3765–3770 (2007).
den Hollander, A.I., Roepman, R., Koenekoop, R.K. & Cremers, F.P. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 27, 391–419 (2008).
Chun, S. & Fay, J.C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Wei, Q., Wang, L., Wang, Q., Kruger, W.D. & Dunbrack, R.L. Jr. Testing computational prediction of missense mutation phenotypes: functional characterization of 204 mutations of human cystathionine β synthase. Proteins 78, 2058–2074 (2010).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B (Methodological) 57, 289–300 (1995).
Liu, Q., Zuo, J. & Pierce, E.A. The retinitis pigmentosa 1 protein is a photoreceptor microtubule-associated protein. J. Neurosci. 24, 6427–6436 (2004).
Davis, E.E. et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 43, 189–196 (2011).
Acknowledgements
We thank M. Sousa, D. Harnley, M.-E. Lancelot and A. Antonio for their excellent technical assistance, The Children's Hospital of Philadelphia CytoGenomics Laboratory for assistance with the establishment of the fibroblast cell lines and tissue culture and J. Baur for his helpful discussions on NAD+ metabolism. We are grateful to the individuals with LCA and their relatives for their participation in this study.
This work was supported by grants from the US National Institutes of Health (RO1-EY12910 (E.A.P.), R03-DK082446 (M.J.F.), R01-GM097409 (E.N.-O.), P30HD026979 (M.J.F. and R.X.) and P30EY014104 (Massachusetts Eye and Ear Infirmary core support)); the Foundation Fighting Blindness USA (I.A., A.D.B., E.L.B., S.S.B., Q.L., A.T.M., D.S.M., E.A.P., J.-A.S., S.M.-S. and A.R.W.); the Rosanne Silbermann Foundation (E.A.P.); the Penn Genome Frontiers Institute (E.A.P. and X.G.); the Institutional Fund to the Center for Biomedical Informatics by the Loyola University Stritch School of Medicine (X.G.); the Foerderer Award for Excellence from the Children's Hospital of Philadelphia (M.J.F. and X.G.); the Angelina Foundation Fund from the Division of Child Development and Metabolic Disease at the Children's Hospital of Philadelphia (M.J.F.); the Clinical and Translational Research Center at the Children's Hospital of Philadelphia (UL1-RR-024134) (M.J.F. and E.A.P.); the Department of Biotechnology, the Government of India and the Champalimaud Foundation, Portugal (C.K.); the Hyderabad Eye Research Foundation (C.K.); a senior research fellowship from the Council for Scientific and Industrial Research (R.S.); the Foundation Voir et Entendre, Ville de Paris and Région Ille de France (C.Z.); RP Fighting Blindness (UK)(A.R.W.); Fight For Sight (UK) (A.D.B., S.S.B., A.T.M., D.S.M. and A.R.W.); Moorfields Eye Hospital National Institute of Health Research (NIHR) British Research Council (BRC) for Ophthalmology (A.D.B., S.S.B., A.T.M., D.S.M. and A.R.W.); and the Special Trustees of Moorfields Eye Hospital (A.D.B., S.S.B., A.T.M., D.S.M. and A.R.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations or the National Institutes of Health. This project is funded, in part, by the Penn Genome Frontiers Institute under a grant with the Pennsylvania Department of Health, which disclaims responsibility for any analyses, interpretations or conclusions.
Author information
Authors and Affiliations
Contributions
Experiments were designed by Q.Z., M.J.F., X.G. and E.A.P. LCA case samples and controls were provided by I.A., A.D.B., E.L.B., S.S.B., C.K., M.J.F., S.J., A.T.M., E.P., S.M.-S., J.-A.S., A.R.W. and E.A.P. Pedigrees were compiled by A.D.B., C.C., C.K., S.J. and E.P. Individuals III-4, III-5, IV-1, IV-2 and IV-3 were clinically evaluated by M.J.F. and E.A.P. (individuals III-3, III-6 and IV-4 to IV-7 were not clinically evaluated by the authors). Exome sequencing was performed by M.C. Bioinformatics pipeline development was performed by X.G., M.J.F., E.A.P. and M.C. Exome data analyses were performed by J.C.P., X.G., Z.F.-K. and E.A.P. NMNAT1 sequencing and segregation analyses were performed by I.A., C.C., M.C., Z.F.-K., D.S.M., L.P., R.S., N.H.W., C.Z. and Q.Z. High-performance liquid chromatography (HPLC)-based NMNAT enzyme activity assay and NAD+ content assay development was performed by E.N.-O., with data analysis performed by E.N.-O., M.J.F., E.A.P. and R.X. In vitro functional studies were carried out by Q.Z., E.N.-O., J.O., Q.L. and M.S. The manuscript was written by M.J.F., Q.Z., X.G. and E.A.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary Information
Rights and permissions
About this article
Cite this article
Falk, M., Zhang, Q., Nakamaru-Ogiso, E. et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat Genet 44, 1040–1045 (2012). https://doi.org/10.1038/ng.2361
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2361
This article is cited by
-
Molecular background of Leber congenital amaurosis in a Polish cohort of patients—novel variants discovered by NGS
Journal of Applied Genetics (2023)
-
NAD+ metabolism and eye diseases: current status and future directions
Molecular Biology Reports (2023)
-
Clinical features and genetic spectrum of NMNAT1-associated retinal degeneration
Eye (2022)
-
Axon Biology in ALS: Mechanisms of Axon Degeneration and Prospects for Therapy
Neurotherapeutics (2022)
-
Photoreceptor metabolic reprogramming: current understanding and therapeutic implications
Communications Biology (2021)