Abstract
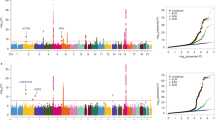
Lung adenocarcinoma is the most common histological type of lung cancer, and its incidence is increasing worldwide. To identify genetic factors influencing risk of lung adenocarcinoma, we conducted a genome-wide association study and two validation studies in the Japanese population comprising a total of 6,029 individuals with lung adenocarcinoma (cases) and 13,535 controls. We confirmed two previously reported risk loci, 5p15.33 (rs2853677, Pcombined = 2.8 × 10−40, odds ratio (OR) = 1.41) and 3q28 (rs10937405, Pcombined = 6.9 × 10−17, OR = 1.25), and identified two new susceptibility loci, 17q24.3 (rs7216064, Pcombined = 7.4 × 10−11, OR = 1.20) and 6p21.3 (rs3817963, Pcombined = 2.7 × 10−10, OR = 1.18). These data provide further evidence supporting a role for genetic susceptibility in the development of lung adenocarcinoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Colvy, T.V. et al. Adenocarcinoma. in World Health Organization Classification of Tumors: Pathology and Genetics, Tumours of Lung, Pleura, Thymus and Heart (eds. Travis, W.D., Brambilla, E., Muller-Hermelink, H.K. & Harris, C.C.) 35–44 (IARC Press, Lyon, France, 2004).
Subramanian, J. & Govindan, R. Lung cancer in never smokers: a review. J. Clin. Oncol. 25, 561–570 (2007).
Sun, S., Schiller, J.H. & Gazdar, A.F. Lung cancer in never smokers—a different disease. Nat. Rev. Cancer 7, 778–790 (2007).
Broderick, P. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 69, 6633–6641 (2009).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40, 1407–1409 (2008).
McKay, J.D. et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 40, 1404–1406 (2008).
Hung, R.J. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452, 633–637 (2008).
Amos, C.I. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 40, 616–622 (2008).
Landi, M.T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Miki, D. et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat. Genet. 42, 893–896 (2010).
Wang, Y. et al. Variation in TP63 is associated with lung adenocarcinoma in the UK population. Cancer Epidemiol. Biomarkers Prev. 20, 1453–1462 (2011).
Hu, Z. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat. Genet. 43, 792–796 (2011).
Freedman, M.L. et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 36, 388–393 (2004).
Yoon, K.A. et al. A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum. Mol. Genet. 19, 4948–4954 (2010).
Ruthenburg, A.J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Medina, P.P. & Sanchez-Cespedes, M. Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer. Epigenetics 3, 64–68 (2008).
Wilson, B.G. & Roberts, C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 (2011).
Hirota, T. et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat. Genet. 43, 893–896 (2011).
Jin, Y. et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N. Engl. J. Med. 362, 1686–1697 (2010).
Asano, K. et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet. 41, 1325–1329 (2009).
Anderson, C.A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43, 246–252 (2011).
Nakamura, Y. The BioBank Japan Project. Clin. Adv. Hematol. Oncol. 5, 696–697 (2007).
Kohno, T. et al. Individuals susceptible to lung adenocarcinoma defined by combined HLA-DQA1 and TERT genotypes. Carcinogenesis 31, 834–841 (2010).
Travis, W.D. et al. World Health Organization. International Histological Classification of Tumors: Histological Typing of Lung and Pleural Tumors (eds. Travis, W.D., Colby, T.V., Corrin, B., Shimosato, Y. & Brambilla, E.) (Springer-Verlag, Heidelberg, Germany, 1999).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Yamaguchi-Kabata, Y. et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am. J. Hum. Genet. 83, 445–456 (2008).
Breslow, N.E. & Day, N.E. Statistical Methods in Cancer Research. Volume II—The Design and Analysis of Cohort Studies 2–333 (IARC Scientific Publications, Lyon, France, 1987).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Acknowledgements
We thank all of the subjects for participating in the study, and we also thank the collaborating physicians for assisting with sample collection. We are grateful to the members of BioBank Japan, the National Cancer Center Biobank and the Rotary Club of Osaka-Midosuji District 2660 Rotary International in Japan for supporting our study. We thank Y. Aoi, T. Odaka, M. Okuyama, H. Totsuka, S. Chiku, A. Kuchiba and the technical staff of the Center for Genome Medicine, National Cancer Center Research Institute, for providing technical and methodological assistance. We also thank H. Hirose of Health Center, Keio University and D. Saito of National Cancer Center Hospital (present affiliation: Nihonbashi Daizo Clinic) for DNA samples of control subjects. This work was supported in part by Grants-in-Aid from the Ministry of Health, Labor and Welfare for Research on Applying Health Technology and for the 3rd-term Comprehensive 10-year Strategy for Cancer Control; from the Ministry of Education, Culture, Sports, Science and Technology of Japan for Scientific Research on Innovative Areas (22131006); from the Japan Society for the Promotion of Science for Research Activity Start-up (23800073) and for Young Scientists (B) (24790340); and by the National Cancer Center Research and Development Fund. This work was also conducted as a part of the BioBank Japan Project supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan. The National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund, Japan.
Author information
Authors and Affiliations
Contributions
K.S., J.Y., M.K. and T.K. designed the study. A.T., K.A., S.O., N.K. and A.S. analyzed the GWAS and replication study. H.S., Y.S., T.Y. and K.S. performed the genotyping for the GWAS and the replication study. H.K., K.G., S.W. and K.T. recruited subjects and participated in diagnostic evaluations. K.S. and T.K. wrote the manuscript. M.K., Y.D., T.Y. and Y.N. contributed to the overall GWAS design.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1 and 2 and Supplementary Tables 1–8. (PDF 2431 kb)
Rights and permissions
About this article
Cite this article
Shiraishi, K., Kunitoh, H., Daigo, Y. et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet 44, 900–903 (2012). https://doi.org/10.1038/ng.2353
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2353
This article is cited by
-
Contribution of an Asian-prevalent HLA haplotype to the risk of HBV-related hepatocellular carcinoma
Scientific Reports (2023)
-
Ethnicity-specific association between TERT rs2736100 (A > C) polymorphism and lung cancer risk: a comprehensive meta-analysis
Scientific Reports (2023)
-
Genome-wide association study of lung adenocarcinoma in East Asia and comparison with a European population
Nature Communications (2023)
-
Comprehensive analysis reveals the potential value of inflammatory response genes in the prognosis, immunity, and drug sensitivity of lung adenocarcinoma
BMC Medical Genomics (2022)
-
Cancer cell-expressed BTNL2 facilitates tumour immune escape via engagement with IL-17A-producing γδ T cells
Nature Communications (2022)