Abstract
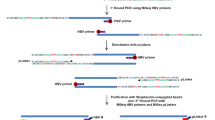
To survey hepatitis B virus (HBV) integration in liver cancer genomes, we conducted massively parallel sequencing of 81 HBV-positive and 7 HBV-negative hepatocellular carcinomas (HCCs) and adjacent normal tissues. We found that HBV integration is observed more frequently in the tumors (86.4%) than in adjacent liver tissues (30.7%). Copy-number variations (CNVs) were significantly increased at HBV breakpoint locations where chromosomal instability was likely induced. Approximately 40% of HBV breakpoints within the HBV genome were located within a 1,800-bp region where the viral enhancer, X gene and core gene are located. We also identified recurrent HBV integration events (in ≥4 HCCs) that were validated by RNA sequencing (RNA-seq) and Sanger sequencing at the known and putative cancer-related TERT, MLL4 and CCNE1 genes, which showed upregulated gene expression in tumor versus normal tissue. We also report evidence that suggests that the number of HBV integrations is associated with patient survival.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Center, M.M. & Jemal, A. International trends in liver cancer incidence rates. Cancer Epidemiol. Biomarkers Prev. 20, 2362–2368 (2011).
But, D.Y., Lai, C.L. & Yuen, M.F. Natural history of hepatitis-related hepatocellular carcinoma. World J. Gastroenterol. 14, 1652–1656 (2008).
Ishikawa, T. Clinical features of hepatitis B virus–related hepatocellular carcinoma. World J. Gastroenterol. 16, 2463–2467 (2010).
Bréchot, C., Gozuacik, D., Murakami, Y. & Paterlini-Brechot, P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin. Cancer Biol. 10, 211–231 (2000).
Zucman-Rossi, J. & Laurent-Puig, P. Genetic diversity of hepatocellular carcinomas and its potential impact on targeted therapies. Pharmacogenomics 8, 997–1003 (2007).
Gehring, A.J. et al. Profile of tumor antigen–specific CD8 T cells in patients with hepatitis B virus–related hepatocellular carcinoma. Gastroenterology 137, 682–690 (2009).
Shafritz, D.A., Shouval, D., Sherman, H.I., Hadziyannis, S.J. & Kew, M.C. Integration of hepatitis B virus DNA into the genome of liver cells in chronic liver disease and hepatocellular carcinoma. Studies in percutaneous liver biopsies and post-mortem tissue specimens. N. Engl. J. Med. 305, 1067–1073 (1981).
Koshy, R. et al. Integration of hepatitis B virus DNA: evidence for integration in the single-stranded gap. Cell 34, 215–223 (1983).
Brechot, C., Pourcel, C., Louise, A., Rain, B. & Tiollais, P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature 286, 533–535 (1980).
Chakraborty, P.R., Ruiz-Opazo, N., Shouval, D. & Shafritz, D.A. Identification of integrated hepatitis B virus DNA and expression of viral RNA in an HBsAg-producing human hepatocellular carcinoma cell line. Nature 286, 531–533 (1980).
Bonilla Guerrero, R. & Roberts, L.R. The role of hepatitis B virus integrations in the pathogenesis of human hepatocellular carcinoma. J. Hepatol. 42, 760–777 (2005).
Murakami, Y. et al. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 54, 1162–1168 (2005).
Paterlini-Bréchot, P. et al. Hepatitis B virus–related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 22, 3911–3916 (2003).
Gozuacik, D. et al. Identification of human cancer-related genes by naturally occurring Hepatitis B Virus DNA tagging. Oncogene 20, 6233–6240 (2001).
Saigo, K. et al. Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma. Hum. Mutat. 29, 703–708 (2008).
Ferber, M.J. et al. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene 22, 3813–3820 (2003).
Neuveut, C., Wei, Y. & Buendia, M.A. Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 52, 594–604 (2010).
Totoki, Y. et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 43, 464–469 (2011).
Li, M. et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011).
Furuta, T. et al. Clinicopathologic features of hepatocellular carcinoma in young patients. Cancer 66, 2395–2398 (1990).
Kim, J.H. et al. Clinical features and prognosis of hepatocellular carcinoma in young patients from a hepatitis B–endemic area. J. Gastroenterol. Hepatol. 21, 588–594 (2006).
Hao, K. et al. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer 9, 389 (2009).
Xu, X. et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29, 735–741 (2011).
Chen, Y. et al. Complete genome sequence of hepatitis B virus (HBV) from a patient with fulminant hepatitis without precore and core promoter mutations: comparison with HBV from a patient with acute hepatitis infected from the same infectious source. J. Hepatol. 38, 84–90 (2003).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Lamb, J.R. et al. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PLoS ONE 6, e20090 (2011).
Chiang, D.Y. et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods 6, 99–103 (2009).
Mermel, C.H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Acknowledgements
We gratefully acknowledge Y.-K. Mak and the clinical team of the Division of Hepatobiliary and Pancreatic Surgery (HBP) at Queen Mary Hospital. This study was funded by the Asian Cancer Research Group (ACRG), a not-for-profit organization formed by Eli Lilly, Merck and Pfizer. We thank S. Friend and G. Jin for initiating the establishment of ACRG. We are grateful to former and present members of ACRG, especially K. Blanchard, Y. Turpaz, J. Sedgwick, G. Tucker-Kellogg, G. Gilliland, P. Shaw, N. Gibson and S. Adams.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Table 1 and Supplementary Note (PDF 1422 kb)
Supplementary Table 2
Summary of sequencing depth and coverage of 88 (TU/AN) pairs of HCC. (XLS 46 kb)
Supplementary Table 3
Characterization of 399 HBV integration breakpoints identified in HCC. (XLS 139 kb)
Supplementary Table 4
Validation result for the HBV integration sites. (XLS 40 kb)
Supplementary Table 5
Correlation analysis of HBV integration (cutoff = 1 to 6) with various clinic-pathological parameters of HCC patients (n = 83). (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Sung, WK., Zheng, H., Li, S. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 44, 765–769 (2012). https://doi.org/10.1038/ng.2295
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2295
This article is cited by
-
Therapeutic efficacy of sorafenib and plant-derived phytochemicals in human colorectal cancer cells
BMC Complementary Medicine and Therapies (2023)
-
Non-canonical MLL1 activity regulates centromeric phase separation and genome stability
Nature Cell Biology (2023)
-
Characteristics of Hepatitis B virus integration and mechanism of inducing chromosome translocation
npj Genomic Medicine (2023)
-
Global burden of hepatitis B virus: current status, missed opportunities and a call for action
Nature Reviews Gastroenterology & Hepatology (2023)
-
Mitochondrial DNA is a target of HBV integration
Communications Biology (2023)