Abstract
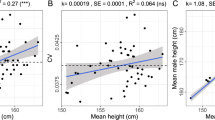
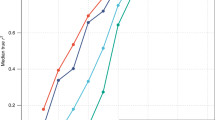
Adult height is a model polygenic trait, but there has been limited success in identifying the genes underlying its normal variation. To identify genetic variants influencing adult human height, we used genome-wide association data from 13,665 individuals and genotyped 39 variants in an additional 16,482 samples. We identified 20 variants associated with adult height (P < 5 × 10−7, with 10 reaching P < 1 × 10−10). Combined, the 20 SNPs explain ∼3% of height variation, with a ∼5 cm difference between the 6.2% of people with 17 or fewer 'tall' alleles compared to the 5.5% with 27 or more 'tall' alleles. The loci we identified implicate genes in Hedgehog signaling (IHH, HHIP, PTCH1), extracellular matrix (EFEMP1, ADAMTSL3, ACAN) and cancer (CDK6, HMGA2, DLEU7) pathways, and provide new insights into human growth and developmental processes. Finally, our results provide insights into the genetic architecture of a classic quantitative trait.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Macgregor, S., Cornes, B.K., Martin, N.G. & Visscher, P.M. Bias, precision and heritability of self-reported and clinically measured height in Australian twins. Hum. Genet. 120, 571–580 (2006).
Preece, M.A. The genetic contribution to stature. Horm. Res. 45, 56–58 (1996).
Silventoinen, K., Kaprio, J., Lahelma, E. & Koskenvuo, M. Relative effect of genetic and environmental factors on body height: differences across birth cohorts among Finnish men and women. Am. J. Public Health 90, 627–630 (2000).
Silventoinen, K. et al. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res. 6, 399–408 (2003).
Perola, M. et al. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 3, e97 (2007).
Palmert, M.R. & Hirschhorn, J.N. Genetic approaches to stature, pubertal timing, and other complex traits. Mol. Genet. Metab. 80, 1–10 (2003).
Weedon, M.N. et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 39, 1245–1250 (2007).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Freedman, M.L. et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 36, 388–393 (2004).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Dixon, A.L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007).
Malumbres, M. & Barbacid, M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30, 630–641 (2005).
Southam, L. et al. An SNP in the 5′-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum. Mol. Genet. 16, 2226–2232 (2007).
Miyamoto, Y. et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 39, 529–533 (2007).
Lohmueller, K.E., Pearce, C.L., Pike, M., Lander, E.S. & Hirschhorn, J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 33, 177–182 (2003).
Fisher, R.A. The correlation between relatives on the supposition of Mendelian inheritance. Philosoph. Trans. Royal Soc. Edinburgh 52, 399–433 (1918).
Lettre, G. et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. advance online publication, doi:10.1038/ng.125 (6 April 2008).
Diabetes Genetics Initiative of Broad Institute of Harvard and M.I.T. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007).
Zeggini, E. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–1341 (2007).
Knight, B., Shields, B.M. & Hattersley, A.T. The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr. Perinat. Epidemiol. 20, 172–179 (2006).
Caulfield, M. et al. Genome-wide mapping of human loci for essential hypertension. Lancet 361, 2118–2123 (2003).
Marques-Vidal, P. et al. Prevalence and characteristics of vitamin or dietary supplement users in Lausanne, Switzerland: the CoLaus study. Eur. J. Clin. Nutr. advance online publication, doi: 10.1038/sj.ejcn.1602932 (17 October 2007).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Stirling, W.D. Enhancements to aid interpretation of probability plots. Statistician 31, 211–220 (1982).
Gauderman, W.J. Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol. 155, 478–484 (2002).
Sandhu, M.S. et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet 371, 483–491 (2008).
Acknowledgements
M.N.W. is a Vandervell Foundation Research Fellow. C.L. is a Nuffield Department of Medicine Scientific Leadership Fellow. R.M.F. is funded by a Diabetes UK research studentship. S.B. is supported by the Giorgi-Cavaglieri Foundation and the Swiss National Science Foundation (grant 3100AO-116323/1), which also supports J.S.B. (grant 310000-112552/1). We would like to thank M. Bochud, Z. Kutalik, G. Waeber, K. Song and X. Yuan for their contribution to the Lausanne study. The WTCCC CAD cohort collection was supported by grants from the British Heart Foundation, Medical Research Council and National Health Service Research & Development. N.J.S. holds a chair supported by the British Heart Foundation. We thank the Wellcome Trust for funding. C.W. is funded by the British Heart Foundation (grant number FS/05/061/19501). The BRIGHT study is supported by the Medical Research Council (grant number G9521010D) and the British Heart Foundation (grant number PG02/128).
Author information
Authors and Affiliations
Consortia
Contributions
M.N.W., H.L., C.M.L., C.W., D.M.E., M.M., J.R.B.P., S.S., I.P., members of the DGI, WTCCC, the GEM consortium, S.B., T.J. and D.M.W. were responsible for analyzing, quality control checking and cleaning the data from the individual GWA studies. C.W., R.M.F., B.S., M.N.W. and H.L. were responsible for analysis of the stage 2 samples. M.N.W. performed the meta-analyses. A.S.H. and N.J.S. are principal investigators from the WTCCC-CAD study. M.C. and M.F. are principal investigators from the WTCCC-HT study. W.H.O. is principal investigator of the WTCCC-UKBS study. A.T.H. and M.I.M. are principal investigators for the WTCCC-T2D study. J.S.B., P.V. and V.M. are principal investigators of the CoLaus study. M.C., M.F., A.D. and P.B.M. are principal investigators on the BRIGHT study. A.T.H. is principal investigator of the EFSOCH study. C.N.A.P. and A.D.M. are principal investigators of the Tayside UKT2D-GCC study. M.N.W., H.L., A.T.H., M.I.M. and T.M.F. wrote the manuscript. A.T.H., M.I.M., M.N.W. and T.M.F. designed and led the study. All authors read and approved the final manuscript.
Corresponding author
Additional information
A full list of authors is provided in the Supplementary Note
A full list of authors is provided in the Supplementary Note
A full list of authors and affiliations appears at the end of this paper.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5, Supplementary Figures 1–3, Supplementary Note (PDF 4393 kb)
Rights and permissions
About this article
Cite this article
Weedon, M., Lango, H., Lindgren, C. et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet 40, 575–583 (2008). https://doi.org/10.1038/ng.121
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.121
This article is cited by
-
Protein-altering variants at copy number-variable regions influence diverse human phenotypes
Nature Genetics (2024)
-
Patterns of enrichment and acceleration in evolutionary rates of promoters suggest a role of regulatory regions in cetacean gigantism
BMC Ecology and Evolution (2023)
-
Genomic signatures of selection, local adaptation and production type characterisation of East Adriatic sheep breeds
Journal of Animal Science and Biotechnology (2023)
-
The molecular evolution of genes previously associated with large sizes reveals possible pathways to cetacean gigantism
Scientific Reports (2023)
-
Global dispersal and adaptive evolution of domestic cattle: a genomic perspective
Stress Biology (2023)