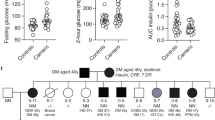
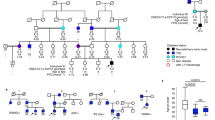
Abstract
Understanding the regulation of pancreatic development is key for efforts to develop new regenerative therapeutic approaches for diabetes. Rare mutations in PDX1 and PTF1A can cause pancreatic agenesis, however, most instances of this disorder are of unknown origin. We report de novo heterozygous inactivating mutations in GATA6 in 15/27 (56%) individuals with pancreatic agenesis. These findings define the most common cause of human pancreatic agenesis and establish a key role for the transcription factor GATA6 in human pancreatic development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Sellick, G.S. et al. Nat. Genet. 36, 1301–1305 (2004).
Stoffers, D.A. et al. Nat. Genet. 15, 106–110 (1997).
Thomas, I.H. et al. Pediatr. Diabetes 10, 492–496 (2009).
Balasubramanian, M. et al. Am. J. Med. Genet. A. 152A, 340–346 (2010).
Kodo, K. et al. Proc. Natl. Acad. Sci. USA 106, 13933–13938 (2009).
Lin, X. et al. J. Hum. Genet. 55, 662–667 (2010).
Maitra, M. et al. Pediatr. Res. 68, 281–285 (2010).
Morrisey, E.E. et al. Genes Dev. 12, 3579–3590 (1998).
Watt, A.J. et al. BMC Dev. Biol. 7, 37 (2007).
Decker, K. et al. Dev. Biol. 298, 415–429 (2006).
Rubio-Cabezas, O. et al. Diabetes 60, 1349–1353 (2011).
Rubio-Cabezas, O. et al. Diabetes 59, 2326–2331 (2010).
Servitja, J.M. & Ferrer, J. Diabetologia 47, 597–613 (2004).
Zhou, Q. et al. Nature 455, 627–632 (2008).
Kroon, E. et al. Nat. Biotechnol. 26, 443–452 (2008).
Acknowledgements
The authors thank the families who participated in this study. We are grateful to A. Damhuis, A. Moorhouse and K. Paszkiewicz for their expert technical assistance. We thank H. Yamagishi (Keio University, Japan) for providing the GATA6 plasmid. S.E.F. was the Sir Graham Wilkins, Peninsula Medical School Research Fellow. S.E. and A.T.H. are employed as core members of staff within the National Institute for Health Research–funded Peninsula Clinical Research Facility. The research leading to these results received funding from Diabetes UK, the Wellcome Research Leave Award for Clinical Academics (ref 067463/Z/2/Z) and the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement number 223211 (Collaborative European Effort to Develop Diabetes Diagnostics, CEED3) and grant agreement number FP7-PEOPLE-ITN-2008 (Marie Curie Initial Training Networks, Biology of Liver and Pancreatic Development and Disease).
Author information
Authors and Affiliations
Consortia
Contributions
S.E., S.E.F., J.F. and A.T.H. designed the study. R.C. performed the exome sequencing and the structural modeling. H.L.A. did the bioinformatic analyses. E.D.F. and S.E.F. did the Sanger sequencing analysis and the interpretation of the resulting data. C.S.-S. and A.T.H. analyzed the clinical data. I.A. and J.F. performed the functional studies. H.L.A., C.S.-S., J.F., A.T.H. and S.E. prepared the draft manuscript. All authors contributed to the discussion of the results and the manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Methods, Supplementary Figures 1–3 and Supplementary Tables 1–4. (PDF 399 kb)
Rights and permissions
About this article
Cite this article
Allen, H., Flanagan, S., Shaw-Smith, C. et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet 44, 20–22 (2012). https://doi.org/10.1038/ng.1035
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.1035
This article is cited by
-
Novel pathogenic GATA6 variant associated with congenital heart disease, diabetes mellitus and necrotizing enterocolitis
Pediatric Research (2024)
-
Monogenic Diabetes with GATA6 Mutations: Characterization of a Novel Family and a Comprehensive Analysis of the GATA6 Clinical and Genetics Traits
Molecular Biotechnology (2024)
-
Diagnostik, Therapie und Verlaufskontrolle des Diabetes mellitus im Kindes- und Jugendalter
Die Diabetologie (2023)
-
Diagnostik, Therapie und Verlaufskontrolle des Diabetes mellitus im Kindes- und Jugendalter
Die Diabetologie (2022)
-
Genetics of diaphragmatic hernia
European Journal of Human Genetics (2021)