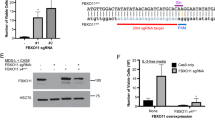
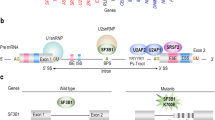
Abstract
Myelodysplastic syndromes (MDS) are hematopoietic stem cell disorders that often progress to chemotherapy-resistant secondary acute myeloid leukemia (sAML). We used whole-genome sequencing to perform an unbiased comprehensive screen to discover the somatic mutations in a sample from an individual with sAML and genotyped the loci containing these mutations in the matched MDS sample. Here we show that a missense mutation affecting the serine at codon 34 (Ser34) in U2AF1 was recurrently present in 13 out of 150 (8.7%) subjects with de novo MDS, and we found suggestive evidence of an increased risk of progression to sAML associated with this mutation. U2AF1 is a U2 auxiliary factor protein that recognizes the AG splice acceptor dinucleotide at the 3′ end of introns, and the alterations in U2AF1 are located in highly conserved zinc fingers of this protein1,2. Mutant U2AF1 promotes enhanced splicing and exon skipping in reporter assays in vitro. This previously unidentified, recurrent mutation in U2AF1 implicates altered pre-mRNA splicing as a potential mechanism for MDS pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Webb, C.J. & Wise, J.A. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol. Cell. Biol. 24, 4229–4240 (2004).
Wu, S., Romfo, C.M., Nilsen, T.W. & Green, M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402, 832–835 (1999).
Welch, J.S. et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. J. Am. Med. Assoc. 305, 1577–1584 (2011).
Mardis, E.R. et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361, 1058–1066 (2009).
Link, D.C. et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. J. Am. Med. Assoc. 305, 1568–1576 (2011).
Ley, T.J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008).
Ley, T.J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010).
Wahl, M.C., Will, C.L. & Luhrmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 (2009).
Fu, Y., Masuda, A., Ito, M., Shinmi, J. & Ohno, K. AG-dependent 3′-splice sites are predisposed to aberrant splicing due to a mutation at the first nucleotide of an exon. Nucleic Acids Res. 39, 4396–4404 (2011).
Kralovicova, J. & Vorechovsky, I. Allele-specific recognition of the 3′ splice site of INS intron 1. Hum. Genet. 128, 383–400 (2010).
Pacheco, T.R., Coelho, M.B., Desterro, J.M., Mollet, I. & Carmo-Fonseca, M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell. Biol. 26, 8183–8190 (2006).
Pacheco, T.R., Moita, L.F., Gomes, A.Q., Hacohen, N. & Carmo-Fonseca, M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol. Biol. Cell 17, 4187–4199 (2006).
Hudson, B.P., Martinez-Yamout, M.A., Dyson, H.J. & Wright, P.E. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11, 257–264 (2004).
Lai, W.S., Kennington, E.A. & Blackshear, P.J. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 277, 9606–9613 (2002).
Liang, J., Song, W., Tromp, G., Kolattukudy, P.E. & Fu, M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE 3, e2880 (2008).
Kielkopf, C.L., Rodionova, N.A., Green, M.R. & Burley, S.K. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106, 595–605 (2001).
Nasim, M.T. & Eperon, I.C. A double-reporter splicing assay for determining splicing efficiency in mammalian cells. Nat. Protoc. 1, 1022–1028 (2006).
Graubert, T.A. et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS ONE 4, e4583 (2009).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Delhommeau, F. et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360, 2289–2301 (2009).
Ebert, B.L. et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451, 335–339 (2008).
Langemeijer, S.M. et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 41, 838–842 (2009).
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42, 722–726 (2010).
Nikoloski, G. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 42, 665–667 (2010).
Gelsi-Boyer, V. et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 145, 788–800 (2009).
Walter, M.J. et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25, 1153–1158 (2011).
Golling, G. et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31, 135–140 (2002).
Rudner, D.Z., Kanaar, R., Breger, K.S. & Rio, D.C. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl. Acad. Sci. USA 93, 10333–10337 (1996).
Zorio, D.A. & Blumenthal, T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5, 487–494 (1999).
Grosso, A.R., Martins, S. & Carmo-Fonseca, M. The emerging role of splicing factors in cancer. EMBO Rep. 9, 1087–1093 (2008).
David, C.J. & Manley, J.L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 (2010).
Visconte, V. et al. SF3B1, a splicing factor, is frequently mutated in refractory anemia with ring sideroblasts. Leukemia published online (2 September 2011), doi:10.1038/leu.2011.232.
Papaemmanuil, E. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Ye, K., Schulz, M.H., Long, Q., Apweiler, R. & Ning, Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Baum, L.E. & Eagon, J.A. An inequality with applications to statistical estimation for probabilistic functions of a Markov process and to a model for ecology. Bull. Am. Math. Soc. 73, 360–363 (1967).
Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6, 677–681 (2009).
Koboldt, D.C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Ng, P.C. & Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003).
Dennis, G. Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Fortier, J.M. et al. POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature. Leukemia 24, 950–957 (2010).
Acknowledgements
This work was supported by US National Institutes of Health grants R01HL082973 (T.A.G.), RC2HL102927 (T.A.G.), U54HG003079 (R.K.W.) and P01CA101937 (T.J.L.) and a Howard Hughes Medical Institute Physician-Scientist Early Career Award (M.J.W.). Technical assistance was provided by the Alvin J. Siteman Cancer Center High Speed Cell Sorting Core, the Molecular and Genomic Analysis Core, the Biomedical Informatics Core and the Tissue Procurement Core, which are all supported by the National Cancer Institute Cancer Center Support Grant P30CA91842. Additional technical assistance was provided by M. Izumi. We thank K. Ohno (Nagoya University Graduate School of Medicine, Japan) for minigene constructs. We thank K. Hall (Washington University School of Medicine) for helpful scientific discussions.
Author information
Authors and Affiliations
Contributions
The project leaders were T.A.G., D.S., L.D. and M.J.W. Study design and project conception were performed by T.A.G., L.D., D.C.L., M.H.T., P.W., J.F.D., E.R.M., T.J.L., R.K.W. and M.J.W. Sequence and data analysis were performed by D.S., L.D., C.C.H., D.C.K., D.E.L., M.D.M., D.J.D., R.M.A., R.S.F., H.S., J.K.-V. and M.O. In vitro splicing assays, PCR or gene expression analyses were performed by T.O.-O., C.L.L., J.S., K.K. and T.N. Clinical data management, specimen acquisition or statistical analyses were performed by M.G., S.H. and J.B. Hematopathology was performed by J.L.F. Manuscript preparation was performed by T.A.G., D.S., L.D., D.C.L., J.F.D., E.R.M., T.J.L., R.K.W. and M.J.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–4 and Supplementary Tables 1–9 (PDF 8748 kb)
Rights and permissions
About this article
Cite this article
Graubert, T., Shen, D., Ding, L. et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 44, 53–57 (2012). https://doi.org/10.1038/ng.1031
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.1031
This article is cited by
-
Impact of U2AF1 mutations on circular RNA expression in myelodysplastic neoplasms
Leukemia (2023)
-
U2AF1 pathogenic variants in myeloid neoplasms and precursor states: distribution of co-mutations and prognostic heterogeneity
Blood Cancer Journal (2023)
-
RNA splicing dysregulation and the hallmarks of cancer
Nature Reviews Cancer (2023)
-
Clinical and prognostic profile of SRSF2 and related spliceosome mutations in patients with acute myeloid leukemia
Molecular Biology Reports (2023)
-
Analysis of genetic variants in myeloproliferative neoplasms using a 22-gene next-generation sequencing panel
BMC Medical Genomics (2022)