Abstract
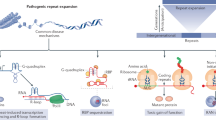
In this Review, familial and sporadic neurological disorders reported to have an etiological link with DNA repair defects are discussed, with special emphasis placed on the molecular link between the disease phenotype and the precise DNA repair defect. Of the 15 neurological disorders listed, some of which have symptoms of progeria, six—spinocerebellar ataxia with axonal neuropathy-1, Huntington's disease, Alzheimer's disease, Parkinson's disease, Down syndrome and amyotrophic lateral sclerosis—seem to result from increased oxidative stress, and the inability of the base excision repair pathway to handle the damage to DNA that this induces. Five of the conditions (xeroderma pigmentosum, Cockayne's syndrome, trichothiodystrophy, Down syndrome, and triple-A syndrome) display a defect in the nucleotide excision repair pathway, four (Huntington's disease, various spinocerebellar ataxias, Friedreich's ataxia and myotonic dystrophy types 1 and 2) exhibit an unusual expansion of repeat sequences in DNA, and four (ataxia-telangiectasia, ataxia-telangiectasia-like disorder, Nijmegen breakage syndrome and Alzheimer's disease) exhibit defects in genes involved in repairing double-strand breaks. The current overall picture indicates that oxidative stress is a major causative factor in genomic instability in the brain, and that the nature of the resulting neurological phenotype depends on the pathway through which the instability is normally repaired.
Key Points
-
The native structure of genomic DNA can be damaged in many ways, both by external agents and from within the cell as a consequence of normal metabolism
-
In an effort to maintain their genomic integrity, organisms have evolved a number of pathways to repair DNA damage
-
Mutations in genes coding for proteins involved in DNA repair pathways can lead to abnormal phenotypes, including neurological disorders, cancer and premature aging; many such conditions are found to have an etiological link to defects in one or more DNA repair pathways
-
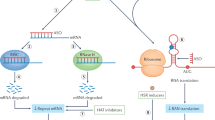
In a postmitotic organ such as the brain, the base excision repair pathway—a conserved mode of DNA repair that deals with the oxidative damage that can result from internal forces—has an important role, and could be a viable candidate for therapeutic targeting
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Rao KS (2003) DNA-repair in brain aging: the importance of base excision repair and DNA polymerase β. Proc Indian Natl Sci Acad B69: 141–156
Reddy MC and Vasquez KM (2005) Repair of genome destabilizing lesions. Radiat Res 164: 345–356
Wood RD et al. (2005) Human DNA repair genes, 2005. Mutat Res 577: 275–283
Wood RD (1996) DNA repair in eukaryotes. Annu Rev Biochem 65: 135–167
Hoeijmakers JHJ (2001) genomic maintenance mechanisms for preventing cancer. Nature 411: 366–374
Mellon I (2005) Transcription-coupled repair: a complex affair. Mutat Res 577: 155–161
Hefferin ML and Tomkinson AE (2005) Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 4: 639–648
Brooks PJ (2002) DNA repair in neural cells: basic science and clinical implications. Mutat Res 509: 93–108
de Boer J and Hoeijmakers JH (2000) Nucleotide excision repair and human syndromes. Carcinogenesis 21: 453–460
Bohr VA et al. (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40: 359–369
Hanawalt PC (1994) Evolution of concepts in DNA repair. Environ Mol Mutagen 23: 78–85
Venema J et al. (1991) Xeroderma pigmentosum complementation group C cells remove pyrimidine dimmers selectively from the transcribed strand of active genes. Mol Cell Biol 11: 4128–4134
Mu D and Sancar A (1997) Model for XPC-independent transcription-coupled repair of pyrimidine dimmers in humans. J Biol Chem 272: 7570–7573
Schofield MJ and Hsieh P (2003) DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol 57: 579–608
Barnes DE and Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38: 445–476
Frosina G et al. (1996) Two pathways for base excision repair in mammalian cells. J Biol Chem 271: 9573–9578
Sokhansanj BA and Wilson DM III (2006) Estimation of the effect of human base excision repair protein variants on the repair of oxidative DNA base damage. Cancer Epidemiol Biomarkers Prev 15: 1000–1008
Dronkert ML and Kanaar R (2001) Repair of DNA interstrand cross-links. Mutat Res 486: 217–247
Thompson LH and Schild D (2002) Recombinational repair and human disease. Mutat Res 509: 49–78
Ren K and de Ortiz SP (2002) Non-homologous DNA end joining in mature rat brain. J Neurochem 80: 949–959
Vyjayanti VN and Rao KS (2006) DNA double strand break repair in brain: reduced NHEJ activity in aging rat neurons. Neurosci Lett 393: 18–22
Rattray AJ and Strathern JN (2003) Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu Rev Genet 37: 31–66
Lehmann AR (2006) Translesion synthesis in mammalian cells. Exp Cell Res 312: 2673–2676
Gratchev A et al. (2003) Molecular genetics of xeroderma pigmentosum variant. Exp Dermatol 12: 529–536
Cleaver JE (2005) Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer 5: 564–573
Groisman R et al. (2006) CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 20: 1429–1434
Fousteri M et al. (2006) Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell 23: 471–482
Graham JM Jr et al. (2001) Cerebro-oculo-facio-skeletal syndrome with a nucleotide excision-repair defect and mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am J Hum Genet 69: 291–300
Andressoo JO et al. (2006) An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria. Cancer Cell 2: 121–132
Raji NS and Rao KS (1998) Trisomy 21 and accelerated aging: DNA-repair parameters in peripheral lymphocytes of Down's syndrome patients. Mech Ageing Dev 100: 85–101
Druzhyna N et al. (1998) Defective repair of oxidative damage in mitochondria DNA in Down's syndrome. Mutat Res 409: 81–89
Fang-Kircher SG et al. (1999) Increases steady state mRNA levels of DNA-repair genes XRCC1, ERCC2 and ERCC3 in brain of patients with Down syndrome. Life Sci 64: 1689–1699
Zana M et al. (2006) Age-dependent oxidative stress-induced DNA damage in Down's lymphocytes. Biochem Biophys Res Commun 345: 726–733
Savitsky K et al. (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268: 1749–1753
Carney JP et al. (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486
Stewart GS et al. (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577–597
Gatei M et al. (2000) ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet 25: 115–119
de Jager M et al. (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell 8: 1129–1135
D'Amours D and Jackson SP (2002) The Mre 11 complex: at the cross-roads of DNA repair and check signaling. Nat Rev Mol Cell Biol 3: 317–327
Lee Y et al. (2001) Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J Neurosci 21: 6687–6693
Biton S et al. (2006) Nuclear ataxia-telangiectasia mutated (ATM) mediates the cellular response to DNA double strand breaks in human neuron-like cells. J Biol Chem 281: 17482–17491
Robinson SH et al. (1987) Alzheimer's disease cells exhibit defective repair of alkylating agent-induced DNA damage. Ann Neurol 21: 250–258
Kodioglu E et al. (2004) Detection of oxidative DNA damage in lymphocytes of patients with Alzheimer's disease. Biomarkers 9: 203–209
Fishel ML et al. (2007) DNA repair in neurons: so if they don't divide what's to repair? Mutat Res 614: 24–36
Shackelford DA (2006) DNA end joining activity is reduced in Alzheimer's disease. Neurobiol Aging 27: 596–605
Fukae J et al. (2005) Expression of 8-oxoguanine DNA glycosylase (OGG1) in Parkinson's disease and related neurodegenerative disorders. Acta Neuropathol (Berl) 109: 256–262
Martin JB (1999) Molecular basis of the neurodegenerative disorders. N Engl J Med 340: 1970–1980
Sieradzan KA and Mann DM (2001) The selective vulnerability of nerve cells in Huntington's disease. Neuropathol Appl Neurobiol 27: 1–21
Li JL et al. (2006) Genome-wide significance for a modifier of age at neurological onset in Huntington disease at 6q23-24: the HD MAPS study. BMC Med Genet 7: 71
Sadri-Vakili G and Cha JH (2006) Mechanisms of disease: histone modifications in Huntington's disease. Nat Clin Pract Neurol 2: 330–338
Hardy J and Orr H (2006) The genetics of neurodegenerative diseases. J Neurochem 97: 1690–1699
Dere R and Wells RD (2006) DM2 CCTG*CAGG repeats are crossover hot spots that are more prone to expansions than the DM1 CTG*CAG repeats in Escherichia coli. J Mol Biol 360: 21–36
El-Khamisy SF et al. (2005) Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434: 108–113
Brooks BP et al. (2005) Genotypic heterogeneity and clinical phenotype in triple A syndrome: a review of the NIH experience 2000–2005. Clin Genet 68: 215–221
Hirano M et al. (2006) ALADINI482S causes selective failure of nuclear protein import and hypersensitivity to oxidative stress in triple A syndrome. Proc Natl Acad Sci USA 103: 2298–2303
Rosen DR et al. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362: 59–62
Petri S et al. (2006) Cell-permeable peptide antioxidants as a novel therapeutic approach in a mouse model of amyotrophic lateral sclerosis. J Neurochem 98: 1141–1148
Kikuchi H et al. (2002) Impairment of mitochondrial DNA repair enzymes against accumulation of 8-oxo-guanine in the spinal motor neurons of amyotrophic lateral sclerosis. Acta Neuropathol (Berl) 103: 408–414
Nagano I et al. (2002) Early decrease of survival factors and DNA repair enzymes in spinal motor neurons of presymptomatic transgenic mice that express a mutant SOD1 gene. Life Sci 72: 541–548
Oh YK et al. (2006) AIF translocates to the nucleus in the spinal motor neurons in a mouse model of ALS. Neurosci Lett 406: 205–210
Garcia-Diaz M et al. (2005) Structure–function studies of DNA polymerase lambda. DNA Repair (Amst) 4: 1358–1367
Rao KS (2006) DNA repair in aging rat neurons. Neuroscience [doi:10.1016/j.neuroscience.2006.09.032]
Acknowledgements
KS Rao would like to acknowledge the help of his colleague Umakanth Swain in drawing the original artwork for figure 1.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Subba Rao, K. Mechanisms of Disease: DNA repair defects and neurological disease. Nat Rev Neurol 3, 162–172 (2007). https://doi.org/10.1038/ncpneuro0448
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ncpneuro0448
This article is cited by
-
Genotoxicity of Psychotropic Drugs in Experimental and Clinical Studies
Neuroscience and Behavioral Physiology (2023)
-
Density functional theory investigations on the interaction of uracil with borospherene
Bulletin of Materials Science (2022)
-
Spontaneous and frequent conformational dynamics induced by A…A mismatch in d(CAA)·d(TAG) duplex
Scientific Reports (2021)
-
PRMT5 promotes DNA repair through methylation of 53BP1 and is regulated by Src-mediated phosphorylation
Communications Biology (2020)
-
AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment
Nature (2020)