Abstract
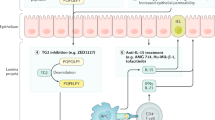
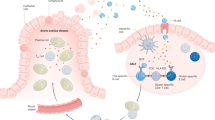
Celiac disease is a disorder of the small intestine caused by an inappropriate immune response to wheat gluten and similar proteins of barley and rye. At present, the only available treatment is a strict gluten-exclusion diet; hence the need for alternative treatments. Recent advances have improved our understanding of the molecular basis for this disorder and there are several attractive targets for new treatments. Oral enzyme supplementation is designed to accelerate gastrointestinal degradation of proline-rich gluten, especially its proteolytically stable antigenic peptides. Complementary strategies aiming to interfere with activation of gluten-reactive T cells include the inhibition of intestinal tissue transglutaminase activity to prevent selective deamidation of gluten peptides, and blocking the binding of gluten peptides to the HLA-DQ2 or HLA-DQ8 molecules. Other possible treatments include cytokine therapy, and selective adhesion molecule inhibitors that interfere with inflammatory reactions, some of which are already showing promise in the clinic for other gastrointestinal diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Mäki M et al. (2003) Prevalence of celiac disease among children in Finland. N Engl J Med 348: 2517–2524
Sollid LM (2002) Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol 2: 647–655
Hausch F et al. (2002) Intestinal digestive resistance of immunodominant gliadin peptides. Am J Physiol Gastrointest Liver Physiol 283: G996–G1003
Vanhoof G et al. (1994) Cloning and sequence analysis of the gene encoding human lymphocyte prolyl endopeptidase. Gene 149: 363–366
Shan L et al. (2002) Structural basis for gluten intolerance in celiac sprue. Science 297: 2275–2279
Piper JL et al. (2004) Effect of prolyl endopeptidase on digestive-resistant gliadin peptides in vivo. J Pharmacol Exp Ther 311: 213–219
Shan L et al. (2004) Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J 383: 311–318
Marti T et al. (2005) Prolyl endopeptidase-mediated destruction of T cell epitopes in whole gluten: chemical and immunological characterization. J Pharmacol Exp Ther 312: 19–26
Chen YS et al. (2003) Identification and characterization of Lactobacillus helveticus PepO2, an endopeptidase with post-proline specificity. Appl Environ Microbiol 69: 1276–1282
Zhang N and Jones BL (1996) Purification and partial characterization of a 31-kDa cysteine endopeptidase from germinated barley. Planta 199: 565–572
Di Cagno R et al. (2002) Proteolysis by sourdough lactic acid bacteria: effects on wheat flour protein fractions and gliadin peptides involved in human cereal intolerance. Appl Environ Microbiol 68: 623–633
Di Cagno R et al. (2004) Sourdough bread made from wheat and nontoxic flours and started with selected lactobacilli is tolerated in celiac sprue patients. Appl Environ Microbiol 70: 1088–1096
Molberg Ø et al. (2001) T cells from celiac disease lesions recognize gliadin epitopes deamidated in situ by endogenous tissue transglutaminase. Eur J Immunol 31: 1317–1323
Vader W et al. (2002) The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology 122: 1729–1737
Arentz-Hansen H et al. (2002) Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology 123: 803–809
De Laurenzi V and Melino G (2001) Gene disruption of tissue transglutaminase. Mol Cell Biol 21: 148–155
Nanda N et al. (2001) Targeted inactivation of Gh/tissue transglutaminase II. J Biol Chem 276: 20673–20678
Szondy Z et al. (2003) Transglutaminase 2−/− mice reveal a phagocytosis-associated crosstalk between macrophages and apoptotic cells. Proc Natl Acad Sci USA 100: 7812–7817
Choi et al. (2005) Chemistry and biology of dihydroisoxazole derivatives: selective inhibitors of human transglutaminase 2. Chem. Biol., in press
Kim CY et al. (2004) Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci USA 101: 4175–4179
Chatenoud L (2003) CD3-specific antibody-induced active tolerance: from bench to bedside. Nat Rev Immunol 3: 123–132
Burkly LC (2001) CD40 pathway blockade as an approach to immunotherapy. Adv Exp Med Biol 489: 135–152
Appel H et al. (2001) Anergy induction by dimeric TCR ligands. J Immunol 166: 5279–5285.
Maurano F et al. (2001) Intranasal administration of one alpha gliadin can downregulate the immune response to whole gliadin in mice. Scand J Immunol 53: 290–295
Salvati VM et al. (2005) Recombinant human IL-10 suppresses gliadin dependent T cell activation in ex vivo cultured celiac intestinal mucosa. Gut 54: 46–53
Mulder CJ et al. (2001) A pilot study of recombinant human interleukin-10 in adults with refractory coeliac disease. Eur J Gastroenterol Hepatol 13: 1183–1188
Nilsen EM et al. (1995) Gluten specific, HLA-DQ restricted T cells from coeliac mucosa produce cytokines with Th1 or Th0 profile dominated by interferon gamma. Gut 37: 766–776
Maiuri L et al. (2003) Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 362: 30–37
Hüe S et al. (2004) A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 21: 367–377
Maiuri L et al. (2000) Interleukin 15 mediates epithelial changes in celiac disease. Gastroenterology 119: 996–1006
Mention JJ et al. (2003) Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology 125: 730–745
Meresse B et al. (2004) Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 21: 357–366
Ogasawara K et al. (2004) NKG2D blockade prevents autoimmune diabetes in NOD mice. Immunity 20: 757–767
Fasano A et al. (2000) Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet 355: 1518–1519
Molberg Ø et al. Mapping of gluten T cell epitopes in the bread wheat ancestors; implications for celiac disease. Gastroenterology, in press
Vader LW et al. (2003) Characterization of cereal toxicity for celiac disease patients based on protein homology in grains. Gastroenterology 125: 1105–1113
Sjöström H et al. (1998) Identification of a gliadin T-cell epitope in coeliac disease: general importance of gliadin deamidation for intestinal T-cell recognition. Scand J Immunol 48: 111–115
Lundin KE et al. (1993) Gliadin-specific, HLA-DQ(α1*0501,β1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J Exp Med 178: 187–196
Molberg Ø et al. (1998) Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med 4: 713–717
van de Wal Y et al. (1998) Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J Immunol 161: 1585–1588
Vader LW et al. (2002) Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J Exp Med 195: 643–649
Forsberg G et al. (2002) Paradoxical coexpression of proinflammatory and down-regulatory cytokines in intestinal T cells in childhood celiac disease. Gastroenterology 123: 667–678
Clemente MG et al. (2003) Early effects of gliadin on enterocyte intracellular signalling involved in intestinal barrier function. Gut 52: 218–223
Colombel JF et al. (2001) Interleukin 10 (Tenovil) in the prevention of postoperative recurrence of Crohn's disease. Gut 49: 42–46
Yasuda H and Wen L (2004) Prevention of autoimmune diabetes by dendritic cells treated with IL-10 [abstract 358-OR]. Diabetes 53
Villadsen LS et al. (2002) HuMax-IL15 reduces the severity of psoriasis in human skin grafts transplanted on to SCID mice. Brit J Dermatol 147: 1064
Ferrari-Lacraz S et al. (2004) Targeting IL-15 receptor-bearing cells with an antagonist mutant IL-15/Fc protein prevents disease development and progression in murine collagen-induced arthritis. J Immunol 173: 5818–5826
Hommes et al. (2004) Fontolizumab (HuZAF), a humanized anti-IFN-gamma antibody, has clinical activity and excellent tolerability in moderate to severe Crohn's disease. Gastroenterology 127: 332
Arulanandam T (2004) Biological characterisitics of anti-alpha4 intergrin monoclonal antibody (natalizumab) a selective adhesion molecule (SAM) inhibitor for the treatment of multiple sclerosis and Crohn's disease [SA23]. Inflamm Res 53: 3
Feagan B et al. (2003) A randomized, double-blind, placebo-controlled trial of iv MLN-02 in 181 patients with moderately active ulcerative colitis is reported. Am J Gastroenterol 98: s248–s249
Fasano A (2002) Peptide antagonists of zonulin and methods for use of the same. US patent 6,548,925
Acknowledgements
Work by the authors was supported in part by grants from the Research Council of Norway, the NIH and the European Commission.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Ludvig M Sollid and Chaitan Khosla are involved in non-profit organizations that develop new treatments for celiac disease. Chaitan Khosla is the President of the Celiac Sprue Research Foundation, and Ludvig M Sollid is a member of the scientific advisory boards of the Celiac Sprue Research Foundation and the National Foundation for Celiac Awareness.
Glossary
- GLUTEN
-
Wheat gluten remains after washing dough and consists of a complex mixture of many gliadin and glutenin polypeptides; gluten-like proteins are also found in rye and barley
- HLA-DQ2 and HLA-DQ8
-
HLA-DQ2 (DQA1*05/DQB1*02) and HLA-DQ8 (DQA1*03/DQB1*0302) are HLA (human leukocyte antigen) molecules associated with celiac disease
- DEAMIDATION
-
The modification of glutamine residues in peptides and proteins to glutamate, or asparagine residues to aspartate
- TISSUE TRANSGLUTAMINASE (TRANSGLUTAMINASE 2; TG2)
-
An enzyme responsible for modifying proteins/peptides by transamidation/deamidation of specific glutamine residues; patients with celiac disease have IgA and IgG antibodies against TG2
- MIC
-
MHC class-I like molecule expressed on gut epithelium; MIC is a ligand for NKG2D
- NKG2D
-
Activating receptor of natural killer cells and CD8+ T cells; NKG2D is expressed on most intraepithelial lymphocytes
- EPITOPE
-
A site on an antigen that is recognized by an antigen receptor (i.e. antibody or T-cell receptor)
- TH1 IMMUNE RESPONSE
-
A type of CD4+ T-cell immune response characterized primarily by the production of interferon-γ
- mutIL15-Fc
-
A lytic and antagonistic IL-15 mutant/Fcgamma2a fusion protein that targets interleukin-15 receptor
- INTEGRIN
-
A cell-surface protein that is involved in cell–cell and cell–matrix interactions
- NOD MICE
-
A mouse strain (nonobese diabetic) that spontaneously develops autoimmune diabetes
- ZONULIN
-
A protein for which the gene is not yet cloned that regulates the permeability of the intestine
Rights and permissions
About this article
Cite this article
Sollid, L., Khosla, C. Future therapeutic options for celiac disease. Nat Rev Gastroenterol Hepatol 2, 140–147 (2005). https://doi.org/10.1038/ncpgasthep0111
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ncpgasthep0111
This article is cited by
-
The less genetic diversity of Triticeae A and D genomes, the smaller diversity in γ-prolamins CD-epitopes
Genetic Resources and Crop Evolution (2018)
-
Significant Hydrolysis of Wheat Gliadin by Bacillus tequilensis (10bT/HQ223107): a Pilot Study
Probiotics and Antimicrobial Proteins (2018)
-
S9A Serine Protease Engender Antigenic Gluten Catabolic Competence to the Human Gut Microbe
Indian Journal of Microbiology (2018)
-
Prolyl-specific peptidases for applications in food protein hydrolysis
Applied Microbiology and Biotechnology (2015)
-
Alternative Approaches Towards Gluten-Free Dough Development: Recent Trends
Food Engineering Reviews (2014)