Abstract
Ndd1 activates the Mcm1–Fkh2 transcription factor to transcribe mitotic regulators. The anaphase-promoting complex/cyclosome activated by Cdh1 (APC/CCdh1) mediates the degradation of proteins throughout G1. Here we show that the APC/CCdh1 ubiquitinates Ndd1 and mediates its degradation, and that APC/CCdh1 activity suppresses accumulation of Ndd1 targets. We confirm putative Ndd1 targets and identify novel ones, many of them APC/CCdh1 substrates. The APC/CCdh1 thus regulates these proteins in a dual manner—both pretranscriptionally and post-translationally, forming a multi-layered feedforward loop (FFL). We predict by mathematical modelling and verify experimentally that this FFL introduces a lag between APC/CCdh1 inactivation at the end of G1 and accumulation of genes transcribed by Ndd1 in G2. This regulation generates two classes of APC/CCdh1 substrates, early ones that accumulate in S and late ones that accumulate in G2. Our results show how the dual state APC/CCdh1 activity is converted into multiple outputs by interactions between its substrates.
Similar content being viewed by others
Introduction
The anaphase-promoting complex/cyclosome (APC/C) was discovered almost 20 years ago as the ubiquitin ligase, which mediates the degradation of mitotic cyclins1,2. The APC/C is a very large complex comprising more than a dozen subunits totalling about 1.5 mD, which is highly conserved in all eukaryotes3. The APC/C is activated in metaphase by the adaptor protein Cdc20 (refs 4, 5). The degradation of Pds1/Securin6 and mitotic cyclins mediated by APC/CCdc20 specific ubiquitination is essential for sister chromatid separation and completion of cell division7,8. The APC/CCdc20 degrades multiple additional mitotic proteins in a well-defined order9 and is the target of the mitotic checkpoint that prevents premature entry into mitosis10
Cdc20 activates the APC/C only during the short mitotic window and is replaced by Cdh1 in late mitosis4,11,12. The APC/CCdh1 remains active throughout G1 until the onset of S phase13 and is constitutively active in metazoan cells in G0 as well as in differentiated cells14. The budding yeast APC/CCdh1 has more than 20 known substrates (Table 1), and this might be only the tip of the iceberg. Some of these substrates are also degraded by APC/CCdc20, but many are unique to APC/CCdh1. In spite of this large number of substrates, Cdh1 is not essential in yeast; however, cdh1Δ cells are sensitive to different types of stress and have a longer cell cycle15. In addition to Cdc20 and Cdh1, the APC/C is activated by Ama1, a third, meiosis-specific, activator16,17.
The increasing number of APC/C substrates discovered in recent years led us18, as well as the Solomon lab19,20, to devise systematic screens to map the entire APC/C ‘degradome’. These screens had an initial bioinformatic step to define a group of candidate genes, followed by various experimental approaches to identify and verify putative substrates. While yielding multiple interesting new APC/C substrates, these screens were limited by the initial biofinformatic step to proteins with a specific transcriptional pattern and degradation box18,19,20. We thus set up an unbiased screen designed to probe the entire yeast proteome. During the pilot stage of this screen, we identified Ndd1, an essential protein, which activates the Mcm1–Fkh2 G2-specific transcription factor21,22,23,24. We show here that Ndd1 is an APC/CCdh1 substrate and that the APC/CCdh1 is a major regulator of the cell cycle-specific oscillations of Ndd1. We demonstrate that the activities of Ndd1 and the APC/CCdh1 reciprocally counteract each other. We used these counteracting activities to verify putative, and identify novel, Ndd1 targets. We tested 16 APC/CCdh1 substrates and found nine of them to be transcribed by Ndd1. The observation that the APC/CCdh1 regulates the level of Ndd1, which transcribes many of the APC/CCdh1 substrates, implies a multi-layered feedforward loop (FFL) that involves both transcriptional and post-translational regulations. We use mathematical modelling to simulate the effect of this Ndd1/APC/CCdh1 FFL on the timing of accumulation and level of APC/CCdh1 substrates. The model predicts that Ndd1 degradation delays the accumulation of these proteins in comparison with APC/CCdh1 substrates that are not transcribed by Ndd1. Live-cell imaging confirms these predictions, and shows that substrates regulated by the Ndd1/APC/CCdh1 FFL accumulate in G2, while those only regulated by the APC/CCdh1 accumulate already in early S phase.
Results
Ndd1 degradation is mediated by the APC/CCdh1
We set up a screen to identify novel APC/CCdh1 substrates in budding yeast. We mated the yeast TAP library25 with a strain expressing constitutively active Cdh1m11 from an inducible GAL promoter26. The library was plated out into 96-well plates with Cdh1m11 induced or uninduced in alternating rows. Protein was extracted from the cultures in each well, dot-blotted on nitrocellulose membranes and immunoblotted with anti-TAP antibodies (Fig. 1a). Using this method, we identified the transcription activator Ndd1 (refs 21, 27) as a putative APC/CCdh1 substrate (Fig. 1b). Ndd1 is the activator of the Mcm1–Fkh2 transcription factor complex21,22,23,24. The protein levels of Ndd1 oscillate periodically and it has been previously speculated28 that it might be degraded by the APC/C. It was further recently shown17 that Ndd1 is degraded in meiosis by the APC/CAma1.
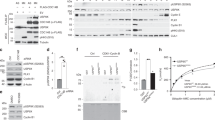
We used several approaches to verify that Ndd1 degradation is indeed mediated by the APC/CCdh1. Figure 2a shows that the endogenous TAP-tagged Ndd1 protein was degraded on activation of Cdh1m11 without reducing the level of its mRNA. Figure 2b shows that Ndd1 accumulated like Clb2 in unsynchronized or mitotic wild-type cells and was virtually absent when these cells were arrested with α factor. In contrast, Ndd1 levels remained high in α factor-arrested cdh1Δ cells.
(a) Levels of endogenous TAP-tagged Ndd1, as well as Clb2, were probed in the absence and presence of Cdh1m11 induction. Tubulin served as a loading control. The level of Ndd1 mRNA was assayed by real-time PCR under both conditions and are shown as the mean of three independent experiments run in duplicates each. Error bars represent s.d. of six samples. (b) Endogenous TAP-tagged Ndd1 was probed in wild-type or cdh1Δ-cycling cells (UT) or cells arrested in prometaphase by Nocodozole (Noc) or in G1 by α factor. Clb2 served as a control for APC/CCdh1 activity and actin as a loading control. FACS analysis of cells harvested in parallel shows that cells were indeed arrested. (c) GST-NDD1 was overexpressed for 2 h by galactose in wild-type and cdh1Δ cells containing the pGAL-NDD1 plasmid and stopped by addition of glucose at time 0. (d) Acm1 expression was induced by galactose in cells harbouring the GAL-ACM1::LEU2 construct. Levels of endogenous Ndd1–Tap and Cdc5 were examined at the indicated time points after Acm1 induction. FACS analysis of cells harvested in parallel shows that Acm1 overexpression does not lead to cell cycle arrest or significantly perturb the cell cycle. (e) Cells expressing HA-tagged wild-type Ndd1 or the Ndd 318KTPA321 (Ndd1KA) putative destruction box mutant were grown in raffinose, Expression was induced for 2 h with galactose and stopped by addition of glucose at time 0. (f) Bacterially expressed GST-Ndd1 was incubated with APC/C and APC/C+Cdh1, resolved by gel electrophoresis and immunoblotted either with anti ubiquitin (left) or anti GST (right) antibodies. The background ubiquitination observed in the left blot in the absence of Ndd1 (right lane) is presumably auto ubiquitination of Cdh1 (ref. 60).
We further used a GAL promoter to induce the expression of GST-tagged Ndd1 in wild-type and in cdh1Δ cells. Once transcription of GST-Ndd1 was turned off by the addition of glucose, Ndd1 was degraded within less than an hour in wild-type cells. In cdh1Δ cells in contrast, Ndd1 levels remained constant for the duration of the experiment (2 h; Fig. 2c).
The activity of the APC/CCdh1 can be inhibited by inducible expression of Acm1 (refs 29, 30, 31). Figure 2d shows that once Acm1 was activated, Ndd1 accumulated at the same rate as that of the known APC/CCdh1 substrate Cdc5. The fact that the Ndd1 levels rise with similar kinetics to those of Cdc5 strongly suggests that its stabilization is a direct consequence of APC/CCdh1 inactivation. If Ndd1 accumulation would have been a downstream event of stabilization of another APC/CCdh1 substrate, its accumulation would have been delayed compared with Cdc5 accumulation. Cdh1Δ cells proliferate relatively normally. We thus did not anticipate that Cdh1 inhibition by Acm1 overexpression would lead to cell cycle arrest. Indeed, fluorescence-activated cell sorting (FACS) analysis showed that Acm1 induction did not significantly perturb the cell cycle (Fig. 2d bottom).
Most APC/CCdh1 substrates require an RxxL32 or KEN33 type ‘destruction box’ to be targeted for ubiquitination and degradation. Ndd1 has five RxxL sequences and no KEN box. According to the GPS-ARM programme34 the 265RTPL268 and the 318RTPL321 rank highest as candidates for a genuine destruction box. We mutated the 318RTPL321 to 318KTPA321 and as shown in Fig. 2e, this mutation significantly stabilized Ndd1. It is rare to obtain complete stabilization by mutagenesis of a single or of even multiple destruction boxes of APC/C substrates in yeast. Indeed, even mutagenesis of all five RxxL sequences did not completely abrogated degradation of Ndd1 by the APC/CAma1 in meiosis17. While all these experiments strongly suggest that Ndd1 is a substrate of the APC/CCdh1, it is not possible to completely exclude the possibility that Ndd1 degradation is an indirect downstream event of APC/CCdh1 activity. We therefore expressed GST-Ndd1 in bacteria and tested its ubiquitination in vitro using APC/C purified from HeLa cells. Figure 2f shows that addition of human Cdh1, expressed by baculovirus and purified from 5B cells to this APC/C, lead to significant monoubiquitination (and more modest polyubiquitination) of Ndd1. We thus conclude that Ndd1 is an APC/CCdh1 ubiquitination substrate.
The APC/CCdh1 regulate Ndd1 levels throughout the cell cycle
Given our observations that Ndd1 is degraded by the APC/CCdh1 in G1, as well as by the APC/CAma1 in meiosis17, we wondered whether Ndd1 is also degraded by the APC/CCdc20 in mitosis. We overexpressed Cdc20 from a GAL promoter in cdh1Δ cells arrested in mitosis with nocodazole. Figure 3a shows that the massive overexpression activated the APC/CCdc20 and led to the degradation of Clb2. Ndd1, on the other hand, remained stable. Ndd1 is thus, like Cdc5, Cdc20 and many other G1-specific APC/CCdh1 substrates, not targeted by the APC/CCdc20.
(a) Cdh1Δ cells expressing gal-inducible Cdc20 were grown to mid-log phase in raffinose. Cells were subsequently arrested by nocodazole for 2 h in mitosis, treated with galactose to induce Cdc20 and sampled at the indicated time points. Cdc20 is as expected strongly induced leading to degradation of Clb2, but not to degradation of Ndd1. Mid-log populations of wild-type (b) or cdh1Δ (c) cells were arrested with alpha factor for two hours, released and sampled at 10-min intervals.
We next sought to follow the level of Ndd1 during the cell cycle and to establish when the APC/CCdh1 is regulating its levels. Ideally, this should have been done in real time in unsynchronized cells. The level of endogenous Ndd1, which is a transcription factor, is however too low to be detected and followed by tagging with a fluorescent protein. We therefore synchronized cells, expressing endogenous TAP-tagged Ndd1 by alpha factor arrest and release. Figure 3b shows that cells start to accumulate Ndd1 20 min after release from alpha factor arrest on initiation of budding. Clb2 accumulation seems to be slightly delayed compared with Ndd1 accumulation, apparently, as will be discussed below, because Ndd1 is required for the transcription of Clb2. Clb2 is degraded on cell division by the APC/CCdc20 and subsequently by APC/CCdh1. The reduction in Ndd1 levels on cell division seems to be slower and less complete, probably due to the fact that it is not degraded by the APC/CCdc20 (Fig. 3a) and also due to loss of synchronization.
The levels of Ndd1 and Clb2 were followed in a similar fashion in cdh1Δ cells (Fig. 3c). Ndd1 levels hardly change in these cells during the cell cycle, indicating that the APC/CCdh1 is the major regulator of Ndd1 levels throughout the cell cycle. As expected, Clb2 levels are higher in cdh1Δ cells and present throughout the entire cell cycle. Clb2 levels still oscillate, presumably due to its degradation by APC/CCdc20 in mitosis.
The activities of Ndd1 and APC/CCdh1 reciprocally counteract
Ndd1 mediates the transcription of mitotic regulators like Clb2 (ref. 27) and Cdc5 (ref. 35), which are degraded by the APC/CCdh1. We thus speculated that Ndd1 could counteract the activity of the APC/CCdh1. To probe the functional interactions between Ndd1 and APC/CCdh1 in vivo, we used GAL-inducible overexpression of Ndd1, as well as of constitutively active Cdh1m11. Figure 4a shows that Ndd1 overexpression impairs growth. Live-cell imaging of these cells shows that they stopped to divide after three to four cycles and extensively grew in size (Fig. 4b). Expression of Cdh1m11 led to cell cycle arrest and to growth of extremely elongated buds (Fig. 4b)36. Strikingly, co-expression of Ndd1 and Cdh1m11 almost eliminated the elongated bud phenotype caused by Cdh1m11 observed after 2 h and greatly reduced it by 24 h (Fig. 4b,c). These observations suggest reciprocal interactions between Ndd1 and the APC/CCdh1—not only does the APC/CCdh1 mediate the degradation of Ndd1 (Fig. 2) but Ndd1 counteracts APC/CCdh1 activity. Interestingly, the inactivation of Ndd1 leads to a cell cycle arrest with elongated buds and replicated DNA27, which is strikingly similar to the phenotype caused by Cdh1m11 expression.
(a) Spot test. 106, 105, 103 and 102 cells with the pGAL-NDD1 plasmid or empty vector were spotted on raffinose and galactose SD uracil-deficient plates and grown for 2 days at 30 °C. (b) Morphology of cells expressing gal-inducible NDD1, Cdh1m11 and both NDD1 and Cdh1m11. Cells were grown to mid-log phase in raffinose, supplemented with galactose and sampled at the indicated time points. (c) The fraction of normal cells, cells with elongated buds and large cells was quantified in duplicates of three independent experiments, n=200 each (1200 in total). Error bars represent s.d. of these three experiments. The length of the scale bar is 10 μm. (d) The indicated strains were subjected to the same experimental conditions as in b and sampled for immunoblotting at the indicated time points.
We hypothesized that the reciprocal effect of Ndd1 and the APC/CCdh1 is caused by the induction of Ndd1-mediated transcription of proteins degraded by the APC/CCdh1. Indeed, Fig. 4d shows that induced expression of Ndd1 rescues the expression of Clb2 and Cdc20, known to be activated by Ndd1 transcriptionally27,37 and degraded by the APC/CCdh1 (ref. 38), even in the presence of constitutively active Cdh1m11.
APC/CCdh1 activation represses Ndd1-specific transcription
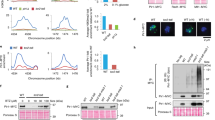
Our observation that the activities of Ndd1 and of the APC/CCdh1 phenotypically counteract (Fig. 4) suggested that APC/CCdh1 activation inhibits the transcription of Ndd1 targets. We thus wondered whether overexpression of Ndd1 and activation of APC/CCdh1 individually and in combination could be used to identify Ndd1 targets. We extracted RNA from cells overexpressing Ndd1, Cdhm11 and Ndd1+Cdhm11 and subjected it to real-time PCR. We initially tested the mRNA levels of genes reported to be transcribed by Ndd1 (CLB1, CLB2, SWI5 (ref. 27), CDC5 (ref. 35) and ACE2 (ref. 39) as well as putative targets whose promoter is bound by Ndd1 (ref. 37; CDC20, SPO12, BUD4 and RAX2). Figure 5a shows that the overexpression of Ndd1 led to significant increase in the mRNA levels of these genes. In contrast, constitutive activation of the APC/CCdh1 led to a significant decrease in their mRNA levels. This decrease is conceivably due to the degradation of the endogenous Ndd1, depleting its activity as a transcriptional activator. Indeed, the combined expression of Ndd1 and Cdh1m11 resulted in an increase of mRNA levels of these genes, but to a lesser extent than when Ndd1 was overexpressed on its own.
Real-time RT–PCR analysis of the indicated genes was performed on RNA extracted from strains overexpressing Ndd1, Cdh1m11 and both. RNA was extracted from samples at time 0 and 1 h. Data are represented as relative expression of each gene at time 0 (taken as 1.0) and are shown as mean of at least two independent experiments performed in duplicates each. Error bars represent s.d. of at least four samples. (a) Analysis of genes previously reported to be transcribed by Ndd1 and of genes whose promoter is bound by Ndd1 (ref. 37). (b) Analysis of genes that are APC/CCdh1 substrates, not previously associated with Ndd1. (c) Analysis of HOF1, member of cluster CLB2 genes that is not an APC/CCdh1 substrate22. (d) Venn diagram of Ndd1 targets, APC/CCdh1 substrates and genes subject to both types of regulation. Refer also to Table 1. RT–PCR, reverse transcription–PCR.
Interestingly, approximately half of these genes are encoding APC/CCdh1 substrates (CLB1, CLB2, CDC5, CDC20 and SPO12), while the rest (RAX2, BUD4, ACE2 and SWI5) are not. We thus expected to find additional novel Ndd1 targets among genes encoding APC/CCdh1 substrates. We tested the effects of overexpression of Ndd1, Cdh1m11 and Ndd1+Cdh1m11 on 11 genes encoding APC/CCdh1 substrates, none of which has previously been associated with Ndd1. Figure 5b shows that four of these genes (ASE1, IQG1, NRM1 and YPH1) were both activated by Ndd1 and repressed by Cdh1m11, as described above, suggesting that they are Ndd1 targets. The mRNA levels of five other genes encoding APC/CCdh1 substrates (CIN8, KIP1, HSL1, PDS1 and RNH202) were neither increased by Ndd1 nor reduced by Cdh1m11, suggesting that they are not transcribed by Ndd1. The mRNA levels of two genes encoding APC/CCdh1 substrates (CIK1 and FIN1) was increased by Ndd1 but not reduced by Cdh1m11. As these two genes did not conform to our stringent dual criterion of activation by Ndd1 and repression by Cdh1m11, we do not define them as Ndd1 targets. It is possible that they are indirect targets or transcribed by Ndd1 as well as by an additional factor. Indeed CIK1 is known to be transcribed by Kar4 (ref. 40), as well as Hcm1 (ref. 41).
Finally, we tested the effect of Ndd1 and Cdh1m11 on mRNA levels of HOF1, a CLB2 cluster gene whose protein is not degraded by the APC/C42. Figure 5c shows that HOF1 mRNA level is strongly induced by Ndd1 overexpression and reduced by Cdh1m11 expression suggesting that HOF1 is transcribed by Ndd1.
Table 1 summarizes the results of these experiments and shows that we identified five novel Ndd1 targets (ASE1, IQG1, NRM1, YPH1 and HOF1), validated four putative ones (CDC20, SPO12, BUD4 and RAX2) and confirmed five previously reported ones (CLB1, CLB2, CDC5, ACE2 and SWI5). Interestingly, the majority of the Ndd1 targets are regulated by the APC/CCdh1, and about half of the APC/CCdh1 substrates we tested are transcribed by Ndd1 (Fig. 5d).
Ndd1 and the APC/CCdh1 generate an FFL
The observation that the APC/CCdh1 exerts a dual effect on some of its substrates, degrading both the proteins themselves and the transcription activator responsible for their synthesis (Fig. 6a), suggests a multi-layered FFL43,44. We hypothesized that APC/CCdh1 substrates subject to the regulation at both levels will have a different pattern of expression compared with APC/CCdh1 substrates regulated only by proteolysis. We reasoned that degradation of the transcription activator mediating transcription of the gene should postpone the reappearance of the translated protein to G2. It should require more time to sequentially synthesize the transcription activator, which subsequently transcribes its target. In contrast, the synthesis of those APC/CCdh1 substrates that are not transcribed by Ndd1 but by another transcription factor could accumulate, depending on the activity of this factor, earlier, already in S phase. We used mathematical modelling and simulations (see Methods) to follow the temporal variation in the levels of shared targets that are regulated by both the APC/CCdh1 and Ndd1 through the wild-type FFL structure (Fig. 6b). These were compared with the target levels obtained when either APC/CCdh1-Ndd1 or APC/CCdh1-target interaction was impaired. The former mimics a situation where the targets are regulated by a transcription factor that is independent of APC/CCdh1. The latter mimics a situation where the targets are substrates of another E3 ligase.
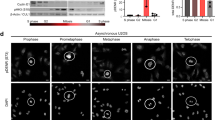
(a) Scheme of an FFL regulatory module generated by APC/CCdh1 and Ndd1. (b) Mathematical simulation predicting the oscillations of protein levels of regulated genes at three conditions, Cdh1 regulates both Ndd1 and target proteins (wild-type regulation, blue line), Ndd1 is not regulated by Cdh1 (turquoise line) and the target protein is not regulated by Cdh1 (magenta line). Arrows point the directions target protein levels are affected by wild-type regulation. (c) Time-lapse series of indicated GFP-labelled proteins in wild-type and cdh1Δ strains, starting with unbudded cells at G1 (t=0). White arrowheads mark GFP-protein accumulation on the bud neck. Asterisk, indicate residual GFP-proteins at G1 stage. The length of the scale bar is 5 μm. See also Supplementary Movies 1, 2, 3, 4 (d) Timing of appearance (GFP on) and disappearance (GFP off) of the indicated GFP-proteins. Time laps series were taken in duplicates in two separate experiments. Error bars represent s.d. of the indicated sample size.
Figure 6b shows the oscillations of a protein regulated dually by Ndd1 and APC/CCdh1 (bottom panel-blue) in relation to APC/CCdh1 activity pattern (top panel). When regulation of Ndd1 by the APC/CCdh1 was abolished, amounting to Ndd1 stabilization, the substrate accumulated earlier (bottom panel- turquoise). On the other hand, when the generic substrate was regulated only by Ndd1 and not by the APC/CCdh1, its accumulation remained late and unchanged (bottom panel-magenta). The timing of its degradation depends on the E3 mediating its ubiquitination. This situation describes the presumed fate of proteins like Hof1 and other proteins transcribed by Ndd1 and not degraded by the APC/CCdh1. The model further predicts that the steady-state levels of substrates dually regulated will be lower than those regulated by either mode independently.
We set out to test the model by following the accumulation of proteins subjected to dual and individual regulations by either APC/CCdh1 or Ndd1, or by both of them together. Fortunately, Iqg1 (APC/CCdh1 and Ndd1), Hsl1 (APC/CCdh1 only) and Hof1 (Ndd1 only) have a highly defined and identical localization to the bud neck facilitating comparison of their accumulation and degradation in real time. We used cells with a carboxyl-terminal GFP (green fluorescent protein)-tagged endogenous gene of each of these three proteins45 and followed asynchronous logarithmic cultures by confocal microscopy. Figure 6c,d shows that the accumulation patterns of these proteins strongly support our mathematical model. Iqg1 (Supplementary Movie 1) regulated by both APC/CCdh1 and Ndd1 accumulates late (44 min after budding). Hsl1 (Supplementary Movie 2), regulated only by the APC/CCdh1 accumulates considerably earlier (16 min after budding). As predicted, both proteins are degraded at exactly the same time (52 min after budding). Hof1 (Supplementary Movie 3), transcribed by Ndd1 and degraded by the SCFGrr1 ubiquitin ligase42 and not by the APC/CCdh1, accumulates, as predicted, at the same time as Iqg1. As Hof1 degradation does not depend on the APC/CCdh1, it takes place later than that of Iqg1 and Hsl1.
Our model further predicted that the elimination of APC/CCdh1 activity would change the time of accumulation of Ndd1 targets. As Iqg1 and Hsl1 are APC/CCdh1 substrates, elimination of APC/CCdh1 leads to their stabilization throughout the cell cycle precluding the possibility to follow their accumulation. However, Hof1 degradation does not depend on APC/CCdh1 and indeed in cdh1Δ cells, where Ndd1 is stabilized, Hof1 accumulates significantly earlier than in wild-type cells (20 min compared with 44 min after budding; Fig. 6c,d and Supplementary Movie 4). Interestingly, this observation matches a report by Blondel et al.42 who replaced the endogenous Hof1 promoter with constitutive CYC1 or ADH promoters and observed that Hof1 started to accumulate shortly after budding, much earlier than when it was expressed from its endogenous promoter. In the experiment we describe here (Fig. 6c bottom panels), we obtained exactly the same effect by deleting CDH1 thereby stabilizing Ndd1. Moreover, as predicted in the model (Fig. 6b turquoise), the level of Hof1 is apparently higher as it is not fully eliminated from previous bud scars (asterisk).
Discussion
The APC/C, which is active from metaphase to the onset of S phase, mediates the ubiquitination and degradation of dozens of proteins. On the background of this crude ‘wholesale’ destruction mechanism, different substrates achieve highly specific temporal patterns of degradation and accumulation. Some of this fine-tuning is achieved via the two activators Cdc20 and Cdh1 that sequentially activate the APC/C46. Recent studies have revealed intricate mechanisms that regulate the temporal pattern of degradation by the APC/CCdc20. Different substrates are degraded at different times, sometimes only minutes apart3,9,47. While the timing of degradation is obviously critical, especially for highly complex events during mitosis, we still know little about what happens at the other end—reaccumulation of these proteins during the S and G2 phases. After an APC/C substrate has been degraded, its accumulation will depend on the time the APC/C is inactivated, as well as on its synthesis. The APC/CCdh1 is turned off at the end of G1 by inactivating phosphorylation of multiple Cdh1 phosphosites26. The inactivation of the APC/CCdh1 at the onset of S phase however does not lead to instantaneous accumulation of all of its substrates, many of which are required only in mitosis. Indeed, most of the substrates regulated by both Ndd1 and the APC/CCdh1 are mitotic proteins. On the other hand, APC/CCdh1 substrates that are not transcribed by Ndd1 may, depending on their respective transcription factor, be expressed either earlier or later. Interestingly, several of these genes are transcribed by the Hcm1 transcription factor41 (Table 1), which is active in S phase before Ndd1 and which also transcribes NDD1.
The FFL generated by the ubiquitin ligase APC/CCdh1, the transcription factor Ndd1, and their shared targets presents an intriguing regulatory circuit. While FFLs comprising two transcription factors were studied previously44, the interaction we describe here is a multi-layered FFL that involves both transcriptional and post-translational regulations. Mathematical simulation of this FFL highlighted two features. The first, which we already discussed, concerns the timing of accumulation of the target proteins in G2 rather than in S. The second effect concerns the extent of substrate repression during G1. Our simulation shows that the steady-state levels of substrates regulated by the FFL are reduced in G1, compared with reduction achieved by any of the regulations on their own. This could be of importance in generating maximal oscillations of activity of these proteins, as well as for achieving effective and rapid repression during G1.
The model in Fig. 6a shows only the basic FFL and has intentionally been stripped of any other interactions. The most relevant interaction is the activating phosphorylation of Ndd1 by Clb1/2 (ref. 48) and Cdc5 (ref. 35). This activation forms in effect an activating positive feedback loop whereby the kinases activate their transcriptional activator.
Ndd1 is degraded during meiosis by the meiotic APC/CAma1 complex and this degradation is required for maintenance of the extended meiotic prophase17. Interestingly, the function of the degradation during the mitotic cycle we describe here seems to be quite different, highlighting how distinct, yet related, ubiquitin ligases can regulate the same protein for diverse purposes during the mitotic and meiotic phases of the cell cycle.
Strikingly, inactivation of Cdh1 eliminates the cell cycle periodicity of the Ndd1 protein (Fig. 3c). This is interesting in view of the periodic transcription by Hcm1 (ref. 41). In the absence of APC/CCdh1 activity, the Ndd1 protein is apparently stable enough so that the periodic transcription has little effect on its level. Cin8, which is also periodically transcribed by Hcm1 (ref. 41) and degraded by the APC/CCdh1 also loses its periodicity on APC/CCdh1 inactivation49.
Ndd1 is the transcriptional activator of Mcm1–Fkh2 in yeast. Mammals, which have a much more complex Forkhead transcription factor network50, lack a direct Ndd1 orthologue. However, in mammals the FoxM1 transcription factor, which transcribes many mitotic genes51 including many APC/CCdh1 substrates, is degraded by the APC/CCdh1 like Ndd1 in budding yeast, and this degradation is critical for entry into S-phase52,53. The apparent evolutionary conservation of this FFL thus confirms its importance for cell cycle regulation.
Methods
Yeast strain construction and growth conditions
Strains used in this study are of S288C background (BY4741, BY4742). Strains with TAP- and GFP-tagged genes were obtained from the corresponding libraries25,45. Yeast strains were engineered by PCR-mediated gene disruption or linearized plasmid integration using standard procedures. Strain cdh1Δ was obtained by replacing CDH1 with KAN. Strains GAL-ACM1 and GAL-CDC20 were obtained by LEU2 integration into its original locus and its disruption by KAN. GAL-ACM1 was integrated into leu2:KAN using plasmid YCplac22-GAL-ACM1-HA31 (a gift from M. Solomon). GAL-CDC20 was integrated into leu2:KAN using plasmid p453-GAL-CDC20 (ref. 54; a gift from F. Uhlmann). Strain GAL-CDH1-m11 was constructed in two steps, disruption of TRP1 by KAN and integration of GAL-CDH1-m11 into trp1:KAN26 (a gift from W. Zachariae). Plasmid pGAL-NDD1-HA [pEG(KG)NDD1] and empty vector [pEG(KG)] both obtained from the MORF collection55 (pBS1805: Gal1p-ORF-6His-HA-3C-ZZ). pGAL-NDD1KA-HA was generated by standard PCR-mediated mutagenesis using the 5′-AACAACATCTTACAGAAGACGCCGGCAAGATCTAACAATAAA-3′ primer.
Yeasts were grown overnight in synthetic defined (SD) medium, diluted 10 × and grown for 2 h to mid-log phase at 30 °C. When proteins were expressed from GAL promoter, cells were grown overnight and diluted in SD raffinose (2%) medium. Protein expression was induced by the addition of galactose (2%). Strains with plasmids were grown at the same conditions but in SD-ura medium.
For obtaining cells at defined cell cycle stages, exponentially growing cells were arrested with 5 μg ml−1 alpha factor (Zymo Research) for 75 min and treated with an additional portion of 5 μg ml−1 alpha factor for 45 min. Cells were either harvested immediately or washed and released into fresh medium and harvested at the indicated time points. Mitotic cells were obtained by treating exponentially growing cells with 15 μg ml−1 Nocodozole (Sigma) for 2 h. Cell samples for FACS analysis were collected simultaneously and processed and analysed as described by Nash et al.56 on a FACSAriaIII (BD Biosciences). For budding, index samples were fixed in 12.5% formaldehyde and washed with double-distilled water before counting. At least 200 cells were counted for each sample.
In vitro ubiquitination assay
Ndd1 was amplified by PCR using 5′-ctatggctagcATGGACAGAGATATAAGCTAC-3′ and 5′-cattctcgagCTGTGAATTGAATAAATTTCC primers (the lowercase letters are overhangs with restriction sites) digested with NheI and XhoI and cloned into pGST-Parallel-3 (ref. 57) digested with SpeI and XhoI. This pGST-NddI expression vector was expressed in bacteria and purified by chromatography on glutathione-sepharose. Recombinant human his6–Cdh1 was expressed in baculovirus-infected 5B insect cells and purified by chromatography on nickel-agarose. APC/C was purified from HeLa cell extract. The ligation of ubiquitin to GST-Ndd1 was assayed in a reaction mixture of 30 μl that contained 30 mM Tris_HCl (pH 7.6), 5 mM MgCl2, 7% glycerol, 1 mM DTT, 1 mg ml−1 bovine serum albumin, 4 mM ATP, 5 mg ml−1 ubiquitin, 0.3 μg of E1, 0.6 μg of E2C/UbcH10, 1 μg of recombinant purified Cdh1, purified APC/C and 0.75 μg GST-Ndd1. The reaction was incubated at 30 °C for 1 h (ref. 58).
Protein extraction and western blotting
To extract protein, yeast cells were fixed with 20% TCA, lysed with glass beads and dissolved in sample buffer15. Protein concentrations were determined using the Coomassie elution with SDS method. Proteins were resolved by SDS–polyacrylamide gel electrophoresis and probed by western blotting. The following antibodies were used: rabbit-α-Cdc5 (Y-300, 1:200), goat-α-Cdc20 (YN-19, 1:100), rabbit-α-Clb2 (Y-180, 1:2000), mouse-α-GST (B-14, 1:100-1:500) all from Santa Cruz, rabbit-α-TAP (CAB101, 1:1000) from Open Biosystems and rabbit-α-ubiquitin (Z045801, 1:5,000), from Dako. Loading controls used were rabbit-α-βactin (ab97376, 1:500; Epitomics) or mouse-α-tubulin (B512, Sigma, 1:10,000). Secondary antibodies were goat-α-rabbit, goat-α-mouse and donkey-α-goat conjugated to HRP from Jackson Laboratories (all 1:10,000).
RNA analysis
Total was RNA extracted using the RNeasy mini kit (Qiagen). One microgram RNA from each sample was used for Oligo (dT)20-primed reverse transcription (Invitrogen) and quantitative reverse transcriptase–PCR reactions were performed using powerSYBR green PCR mix (ABI). Each sample was analysed in duplicates. RNA loading of all samples was normalized for TFC1 levels59. Primers (Supplementary Table 1) were designed using ProbeFinder software, Universal Probe Library for yeast (www.roche-applied-science.com).
Mathematical model
The analyses of the FFL module, composed of Cdh1 that regulates its targets both directly and indirectly via Ndd1, were carried out using a deterministic model, based on rate equations. The model consists of a set of coupled ordinary differential equations, where each equation evaluates the time derivative of the number of one type of molecule.
The rate equations describe the following processes: Cdh1 mRNA (mx) is transcribed at rate  and degraded at rate
and degraded at rate  . Cdh1 protein (Px) is translated at rate
. Cdh1 protein (Px) is translated at rate  and degraded at rate
and degraded at rate  . Cdh1 is transformed into its active form (
. Cdh1 is transformed into its active form ( ) at rate p, and is degraded at rate
) at rate p, and is degraded at rate  . Ndd1 mRNA (my) is transcribed at rate
. Ndd1 mRNA (my) is transcribed at rate  and degraded at rate
and degraded at rate  . Ndd1 protein (Py) is translated at rate
. Ndd1 protein (Py) is translated at rate  and degraded at rate
and degraded at rate  . Ndd1 is bound and degraded by Cdh1 active protein at rate bxy. In addition, Ndd1 binds the promoter of its target gene at rate bz and unbinds the promoter at rate uz. The average number of bound Ndd1 proteins to the target promoter is denoted by Bz. The target mRNA (mz) is transcribed at rate
. Ndd1 is bound and degraded by Cdh1 active protein at rate bxy. In addition, Ndd1 binds the promoter of its target gene at rate bz and unbinds the promoter at rate uz. The average number of bound Ndd1 proteins to the target promoter is denoted by Bz. The target mRNA (mz) is transcribed at rate  , reflecting its transcriptional activation by Ndd1, and it is degraded at rate
, reflecting its transcriptional activation by Ndd1, and it is degraded at rate  . The target protein (Pz) is translated at rate
. The target protein (Pz) is translated at rate  and degraded at rate
and degraded at rate  . The target protein is bound and degraded by Cdh1 active protein at rate bxz. The rate equations describing these processes take the form:
. The target protein is bound and degraded by Cdh1 active protein at rate bxz. The rate equations describing these processes take the form:








This basic FFL module was tweaked in two ways. To reflect a module without Cdh1–Ndd1 regulation, bxy was set to 0. Similarly, in a module without Cdh1-target regulation, bxz=0.
The equations were implemented in MATLAB (MathWorks) and integrated using its built-in solver ode45.
The parameter values used for the simulations are as follows:  ,
,  ,
,  ,
,  , P=0 when Cdh1 is repressed and P=0.001 s−1 when it is activated, bxy=bxz=0.0253 s−1, bz=0.005 s−1, uz=0.05 s−1.
, P=0 when Cdh1 is repressed and P=0.001 s−1 when it is activated, bxy=bxz=0.0253 s−1, bz=0.005 s−1, uz=0.05 s−1.
Live imaging
Cells were plated on conA-coated chambered coverglasses (Mattek) and imaged by an Olympus FV1000 confocal microscope with a 488-nm Argon ion laser equipped with an automated stage, auto-focus device (Z-drift compensation, ZDC) and an environmental chamber (LIS, Switzerland). A × 60 1.4 NA oil immersion objective was used, temperature was maintained at 30 °C and frames were captured every 5 min. The various strains were filmed in parallel in the different wells in the same experiment to ensure identical conditions. Differential interference contrast images were also captured. Data analysis was performed using ImageJ (NIH).
Additional information
How to cite this article: Sajman, J. et al. Degradation of Ndd1 by APC/CCdh1 generates a feedforward loop that times mitotic protein accumulation. Nat. Commun. 6:7075 doi: 10.1038/ncomms8075 (2015).
References
Sudakin, V. et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell 6, 185–197 (1995).
King, R. W. et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81, 279–288 (1995).
Pines, J. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12, 427–438 (2011).
Visintin, R., Prinz, S. & Amon, A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science 278, 460–463 (1997).
Sigrist, S. J., Jacobs, H., Stratmann, R. & Lehner, C. F. Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3. EMBO J. 14, 4827–4838 (1995).
Cohen-Fix, O., Peters, J. M., Kirschner, M. W. & Koshland, D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10, 3081–3093 (1996).
Lim, H. H., Goh, P. Y. & Surana, U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 8, 231–234 (1998).
Shirayama, M., Toth, A., Galova, M. & Nasmyth, K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402, 203–207 (1999).
Lu, D. et al. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J.Cell Biol. 207, 23–39 (2014).
Varetti, G., Guida, C., Santaguida, S., Chiroli, E. & Musacchio, A. Homeostatic control of mitotic arrest. Mol. Cell 44, 710–720 (2011).
Schwab, M., Lutum, A. & Seufert, W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90, 683–693 (1997).
Sigrist, S. J. & Lehner, C. F. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90, 671–681 (1997).
Amon, A., Irniger, S. & Nasmyth, K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell 77, 1037–1050 (1994).
Brandeis, M. & Hunt, T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 15, 5280–5289 (1996).
Simpson-Lavy, K. J., Sajman, J., Zenvirth, D. & Brandeis, M. APC/CCdh1 specific degradation of Hsl1 and Clb2 is required for proper stress responses of S. cerevisiae. Cell Cycle 8, 3003–3009 (2009).
Cooper, K. F., Mallory, M. J., Egeland, D. B., Jarnik, M. & Strich, R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl Acad. Sci. USA 97, 14548–14553 (2000).
Okaz, E. et al. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 151, 603–618 (2012).
Feine, O., Zur, A., Mahbubani, H. & Brandeis, M. Human Kid is degraded by the APC/C(Cdh1) but not by the APC/C(Cdc20). Cell Cycle 6, 2516–2523 (2007).
Ostapenko, D., Burton, J. L. & Solomon, M. J. Identification of anaphase promoting complex substrates in S. cerevisiae. PLoS ONE 7, e45895 (2012).
Ostapenko, D. & Solomon, M. J. Anaphase promoting complex-dependent degradation of transcriptional repressors Nrm1 and Yhp1 in Saccharomyces cerevisiae. Mol. Biol. Cell 22, 2175–2184 (2011).
Koranda, M., Schleiffer, A., Endler, L. & Ammerer, G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406, 94–98 (2000).
Kumar, R. et al. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10, 896–906 (2000).
Zhu, G. et al. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406, 90–94 (2000).
Pic, A. et al. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 19, 3750–3761 (2000).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Zachariae, W., Schwab, M., Nasmyth, K. & Seufert, W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282, 1721–1724 (1998).
Loy, C. J., Lydall, D. & Surana, U. NDD1, a high-dosage suppressor of cdc28-1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 3312–3327 (1999).
Wittenberg, C. & Reed, S. I. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24, 2746–2755 (2005).
Martinez, J. S., Jeong, D. E., Choi, E., Billings, B. M. & Hall, M. C. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol. Cell. Biol. 26, 9162–9176 (2006).
Dial, J. M., Petrotchenko, E. V. & Borchers, C. H. Inhibition of APCCdh1 activity by Cdh1/Acm1/Bmh1 ternary complex formation. J. Biol. Chem. 282, 5237–5248 (2007).
Ostapenko, D., Burton, J. L., Wang, R. & Solomon, M. J. Pseudosubstrate inhibition of the anaphase-promoting complex by Acm1: regulation by proteolysis and Cdc28 phosphorylation. Mol. Cell. Biol. 28, 4653–4664 (2008).
Glotzer, M., Murray, A. W. & Kirschner, M. W. Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138 (1991).
Pfleger, C. M. & Kirschner, M. W. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14, 655–665 (2000).
Liu, Z. et al. GPS-ARM: computational analysis of the APC/C recognition motif by predicting D-boxes and KEN-boxes. PLoS ONE 7, e34370 (2012).
Darieva, Z. et al. Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature 444, 494–498 (2006).
Robbins, J. A. & Cross, F. R. Requirements and reasons for effective inhibition of the anaphase promoting complex activator CDH1. Mol. Biol. Cell 21, 914–925 (2010).
Simon, I. et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106, 697–708 (2001).
Shirayama, M., Zachariae, W., Ciosk, R. & Nasmyth, K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17, 1336–1349 (1998).
Yang, Y. L., Suen, J., Brynildsen, M. P., Galbraith, S. J. & Liao, J. C. Inferring yeast cell cycle regulators and interactions using transcription factor activities. BMC Genomics 6, 90 (2005).
Lahav, R., Gammie, A., Tavazoie, S. & Rose, M. D. Role of transcription factor Kar4 in regulating downstream events in the Saccharomyces cerevisiae pheromone response pathway. Mol. Cell. Biol. 27, 818–829 (2007).
Pramila, T., Wu, W., Miles, S., Noble, W. S. & Breeden, L. L. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 20, 2266–2278 (2006).
Blondel, M. et al. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 24, 1440–1452 (2005).
Milo, R. et al. Network motifs: simple building blocks of complex networks. Science 298, 824–827 (2002).
Mangan, S. & Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11980–11985 (2003).
Huh, W. K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Zur, A. & Brandeis, M. Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J. 21, 4500–4510 (2002).
Sedgwick, G. G. et al. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. EMBO J. 32, 303–314 (2013).
Reynolds, D. et al. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 17, 1789–1802 (2003).
Hildebrandt, E. R. & Hoyt, M. A. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol. Biol. Cell 12, 3402–3416 (2001).
Eijkelenboom, A. & Burgering, B. M. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14, 83–97 (2013).
Fu, Z. et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 10, 1076–1082 (2008).
Park, H. J., Costa, R. H., Lau, L. F., Tyner, A. L. & Raychaudhuri, P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol. Cell Biol. 28, 5162–5171 (2008).
Laoukili, J., Alvarez-Fernandez, M., Stahl, M. & Medema, R. H. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle 7, 2720–2726 (2008).
Lengronne, A. et al. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell 23, 787–799 (2006).
Gelperin, D. M. et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 (2005).
Nash, R., Tokiwa, G., Anand, S., Erickson, K. & Futcher, A. B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7, 4335–4346 (1988).
Sheffield, P., Garrard, S. & Derewenda, Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expr. Purif. 15, 34–39 (1999).
Braunstein, I., Miniowitz, S., Moshe, Y. & Hershko, A. Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc. Natl Acad. Sci. USA 104, 4870–4875 (2007).
Teste, M. A., Duquenne, M., Francois, J. M. & Parrou, J. L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 10, 99 (2009).
Listovsky, T. et al. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 23, 1619–1926 (2004).
Shah, R., Jensen, S., Frenz, L. M., Johnson, A. L. & Johnston, L. H. The Spo12 protein of Saccharomyces cerevisiae: a regulator of mitotic exit whose cell cycle-dependent degradation is mediated by the anaphase-promoting complex. Genetics 159, 965–980 (2001).
Kishi, T., Ikeda, A., Koyama, N., Fukada, J. & Nagao, R. A refined two-hybrid system reveals that SCF(Cdc4)-dependent degradation of Swi5 contributes to the regulatory mechanism of S-phase entry. Proc. Natl Acad. Sci. USA 105, 14497–14502 (2008).
Juang, Y. L. et al. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275, 1311–1314 (1997).
Ko, N. et al. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol. Biol. Cell 18, 5139–5153 (2007).
Gordon, D. M. & Roof, D. M. Degradation of the kinesin Kip1p at anaphase onset is mediated by the anaphase-promoting complex and Cdc20p. Proc. Natl Acad. Sci. USA 98, 12515–12520 (2001).
Burton, J. L. & Solomon, M. J. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol. Cell. Biol. 20, 4614–4625 (2000).
Benanti, J. A., Matyskiela, M. E., Morgan, D. O. & Toczyski, D. P. Functionally distinct isoforms of Cik1 are differentially regulated by APC/C-mediated proteolysis. Mol. Cell 33, 581–590 (2009).
Woodbury, E. L. & Morgan, D. O. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9, 106–112 (2007).
Ferguson, J. L., Chao, W. C., Lee, E. & Friedman, K. L. The anaphase promoting complex contributes to the degradation of the S. cerevisiae telomerase recruitment subunit Est1p. PLoS ONE 8, e55055 (2013).
Ferreira, M. F., Santocanale, C., Drury, L. S. & Diffley, J. F. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 20, 242–248 (2000).
Acknowledgements
We thank Avram Hershko for reagents and help with the in vitro ubiquitination assay of Ndd1, to Tsafi Danieli for help in expressing baculo viral Cdh1, to William Breuer for FACS analysis, to Wolfgang Zachariae, Mark Solomon, Frank Uhlmann and Angelika Amon for yeast strains and to Ofer Biham for fruitful discussions,. This work was funded by grants from the The U.S.-Israel Binational Science Foundation (BSF2007-288-03), the Israel Cancer Foundation (ICRF) and the Israel Science Foundation (1759/14). M.N. is grateful to the Azrieli Foundation for the award of an Azrieli Fellowship.
Author information
Authors and Affiliations
Contributions
J.S., D.Z., Y.R. and M.B. conceived and designed the experiments. J.S., D.Z., Y.R., I.C. and M.B. performed the experiments. M.N. and H.M. performed the mathematical modelling. D.Z., K.J.S.-L., H.M., T.R. and M.B. analysed the data. D.Z. and M.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Table 1. (PDF 95 kb)
Supplementary Movie 1
Time lapse series of Iqg1-GFP cells. (AVI 277 kb)
Supplementary Movie 2
Time lapse series of Hsl1-GFP cells. (AVI 286 kb)
Supplementary Movie 3
Time lapse series of Hof1-GFP cells. (AVI 560 kb)
Supplementary Movie 4
Time lapse series of Hof1-GFP cdh1Δ cells. (AVI 114 kb)
Rights and permissions
About this article
Cite this article
Sajman, J., Zenvirth, D., Nitzan, M. et al. Degradation of Ndd1 by APC/CCdh1 generates a feed forward loop that times mitotic protein accumulation. Nat Commun 6, 7075 (2015). https://doi.org/10.1038/ncomms8075
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms8075
This article is cited by
-
Cyclin/Forkhead-mediated coordination of cyclin waves: an autonomous oscillator rationalizing the quantitative model of Cdk control for budding yeast
npj Systems Biology and Applications (2021)
-
Insights into APC/C: from cellular function to diseases and therapeutics
Cell Division (2016)
-
Create, activate, destroy, repeat: Cdk1 controls proliferation by limiting transcription factor activity
Current Genetics (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.