Abstract
Peptide uptake systems that involve members of the proton-coupled oligopeptide transporter (POT) family are conserved across all organisms. POT proteins have characteristic substrate multispecificity, with which one transporter can recognize as many as 8,400 types of di/tripeptides and certain peptide-like drugs. Here we characterize the substrate multispecificity of Ptr2p, a major peptide transporter of Saccharomyces cerevisiae, using a dipeptide library. The affinities (Ki) of di/tripeptides toward Ptr2p show a wide distribution range from 48 mM to 0.020 mM. This substrate multispecificity indicates that POT family members have an important role in the preferential uptake of vital amino acids. In addition, we successfully establish high performance ligand affinity prediction models (97% accuracy) using our comprehensive dipeptide screening data in conjunction with simple property indices for describing ligand molecules. Our results provide an important clue to the development of highly absorbable peptides and their derivatives including peptide-like drugs.
Similar content being viewed by others
Introduction
Peptide uptake systems are conserved across all organisms from bacteria to higher animals and plants and are important for acquiring nitrogen resources with high efficiency1,2,3,4,5,6. The molecules involved in these systems belong to the proton-coupled oligopeptide transporter (POT) family, which is also called the peptide transporter (PTR) family3. POT family members comprise 12 transmembrane domains as a general structure and have characteristic substrate multispecificity, by which one transporter can recognize a variety of substrates7. In humans, hPEPT1 expressed in the small intestine epithelium is involved in absorbing nutritional peptides8,9,10, whereas hPEPT2 expressed in the renal tubules is involved in the reabsorption of peptides from primitive urine11,12,13.
Due to their substrate multispecificity, it is assumed that each of these POT family transporters can recognize as many as 8,400 types of di/tripeptides, which are products generated by protein hydrolysis, at a single substrate-binding site that resides on each transporter and can actively transport these peptides7,14,15. Furthermore, hPEPT1 and hPEPT2 can transport peptide-like drugs such as β-lactam antibiotics, anti-hypertensive drugs and anti-cancer agents16. This property influences the intestinal absorption of drugs and half-life of drugs in blood. For the yeast S. cerevisiae, Ptr2p is the major transporter involved in the uptake of di/tripeptides17,18,19,20. Because the uptake of amino acids in the peptide form is much faster than in the amino-acid form, peptide-based media can improve the growth of yeasts6,21. Although Ptr2p is an important protein for the fermentation industry, there is little information on the substrate preferences of Ptr2p22. As with the examples above, the substrate multispecificity of a POT family protein is of interest in various fields of science including drug development, nutrition and fermentation.
The question of how one substrate-binding site can recognize a variety of substrates is the major focus in peptide transport research. To date, the necessary or important structural characteristics for recognition by POT family transporters have been proposed by determining the affinity of individual substrates for these transporters23,24,25. However, the entire spectrum of the substrate preference of POT family transporter has not been elucidated. In addition, the crystal structures of two bacterial POT family transporters, PepTso14 from Shewanella oneidensis and PepTst15 from Streptococcus thermophilus, were recently determined, which provided important clues for elucidating the substrate multispecificity of the POT family. Because the amino-acid sequence of PepTso is highly homologous to those of hPEPT1 and hPEPT2, PepTso is the best structural model currently available for these eukaryotic peptide transporters. The crystal structure of PepTso provided basic information on the three dimensional configuration of the amino-acid residues at its substrate-binding site. To advance further in designing a detailed pharmacophore map, it is necessary to elucidate the physicochemical characteristics that determine the affinity of a substrate for a POT family transporter.
This study reports the substrate multispecificity of S. cerevisiae Ptr2p used as a model POT family transporter. Detailed substrate multispecificity of the Ptr2p was characterized by a comprehensive analysis using a dipeptide library and a high-throughput assay system developed by us. By constructing models to predict the dipeptide affinities for Ptr2p, we observed that ligand affinity Ki values could be predicted in silico with high accuracy. For in silico model construction, we used a combination of simple ligand property parameters rather than using complex structural information for ligands and receptors. The biological mechanisms and roles of POT family proteins are discussed on the basis of careful examination of the prediction models constructed using different property parameters as descriptive parameters for ligand affinity.
Results
Construction of a Ptr2p expression system
To avoid the activity of its endogenous peptide transporter, a PTR2 gene knockout strain (S. cerevisiae BY4742-ptr2Δ) was used as the host strain for Ptr2p expression (SC-Ptr2p). The membrane fraction of SC-Ptr2p cells was analysed by western blot using an anti-FLAG antibody to verify Ptr2p expression (Fig. 1a). Ptr2p was detected as a single band of 68 kDa, which was consistent with the size of the intact Ptr2p19. The FLAG tag was replaced with GFP at the carboxy terminus of Ptr2p and localization of the Ptr2p–GFP fusion protein was analysed by confocal fluorescence microscopy (Fig. 1b). The result indicated that Ptr2p–GFP was localized on the cell surface. Dipeptide uptake ability of SC-Ptr2p cells was then analysed (Fig. 1c). Strain BY4742 requires leucine and histidine in its growth medium. Thus, these amino acids were added to the growth medium in the form of dipeptides (His and Leu). SC-Ptr2p cells grew and formed colonies on these plates after absorbing these peptides, whereas its host strain did not grow. These results indicated that Ptr2p was potent, expressed exogenously and localized on the cell surface.
(a) Western blot analysis of the membrane fraction prepared from SC-Ptr2p cells (BY4742-ptr2Δ cells that expressed plasmid-borne Ptr2p) using an anti-FLAG antibody. Arrowhead indicates expressed Ptr2p. (b) Localization of a Ptr2p–GFP fusion protein using confocal fluorescence microscopy in BY4742-ptr2Δ. Scale bar represents 5 μm. (c) Spot assay. Dipeptide uptake capability of SC-Ptr2p cells was analysed. Leucine and histidine, which are required for the growth of strain BY4742 were added to the medium in the form of a dipeptide (His–Leu, 10 mM).
A fluorescence-based competitive uptake assay system
A fluorescence-based competitive uptake (F-CUp) assay system is a high-throughput assay that determines the competitive-inhibitory activity (IC50) of an analysed substrate versus the uptake of a tracer substrate. An IC50 value can be converted to a Ki value using the Cheng–Prusoff equation26. Functional analysis of Ptr2p was conducted using β-Ala–Lys (AMCA) as the tracer substrate to establish an F-CUp assay system. β-Ala–Lys (AMCA) uptake was mediated by Ptr2p and it accumulated within the vacuoles (Fig. 2a). This uptake was inhibited by the imidazole dipeptide carnosine as a representative result. The β-Ala–Lys (AMCA) uptake was time-dependent, and its uptake by the host strain was negligible (Fig. 2b). In previous experiments, it was demonstrated that most of the di/tripeptides were transported via Ptr2p in S. cerevisiae6. Thus, Ptr2p was a major transporter for both the tracer (β-Ala–Lys(AMCA)) and dipeptides. Furthermore, β-Ala–Lys (AMCA) uptake was also concentration dependent, and the Km value was calculated to be 0.16 (±0.02, s.d.) mM according to the Michaelis–Menten formula (Fig. 2c). The competitive-inhibitory activity was analysed using amino acids and oligopeptides with different chain lengths (Fig. 2d). Only di/tripeptides showed competitive-inhibitory effects against the uptake of the tracer substrate. The Ki values for Gly–Gly and Gly–Gly–Gly were calculated to be 17 and 48 mM, respectively. To investigate whether differences in amino-acid sequences affected the affinity for Ptr2p, three different dipeptides, Ala–Ala, Ala–Leu and Leu–Ala, were analysed (Fig. 2e). The affinities of the two dipeptides that included leucine, Leu–Ala (Ki=0.15 mM) and Ala–Leu (Ki=0.31 mM), were higher than that of Ala–Ala (Ki=0.40 mM). The affinity of Leu–Ala was twice that of Ala–Leu. Thus, this demonstrated that in addition to the amino-acid composition, the amino-acid sequence also contributed to the affinity for Ptr2p.
(a) Analysis of tracer substrate, β-Ala–Lys (AMCA), uptake by SC-Ptr2p cells (BY4742-ptr2Δ cells that expressed plasmid-borne Ptr2p) using confocal fluorescence microscopy. Scale bar represents 5 μm. (b) Time course for the uptake of the tracer substrate (50 μM) in SC-Ptr2p. Dark blue: SC-Ptr2p cells, Light blue: parental strain BY4742-ptr2Δ. (c) Concentration dependence of tracer substrate uptake in SC-Ptr2p. Dark blue column: SC-Ptr2p cells; light blue column: parental strain BY4742-ptr2Δ cells. (d) Effect of peptide chain length on tracer substrate uptake based on competitive inhibition. White column: Gly; light green column: Gly–Gly, green column: Gly–Gly–Gly; dark green column: Gly–Gly–Gly–Gly. N.T.: not tested. (e) Effect of amino-acid sequence on tracer substrate uptake based on competitive inhibition. Red column: Ala–Ala, yellow column: Ala–Leu, purple column: Leu–Ala. Results of panels b–e are means±s.d. (n=3).
Relationship between Ptr2p affinity and S. cerevisiae growth
The affinity of dipeptides for Ptr2p and their effects on the growth of S. cerevisiae were examined using two different combinations of dipeptides: His–Leu and Leu-His or Leu-Gly and Gly-Leu (Fig. 3). The Ki values of His–Leu, Leu-His, Leu-Gly and Gly-Leu were 0.05, 0.13, 0.36 and 0.60 mM, respectively. Yeast cell growth analysis indicated that those dipeptides with lower Ki values were better nutrients for Ptr2p-expressing yeast despite their identities in terms of their amino-acid composition. The effect for improving cell growth by a high-affinity peptide was also verified using the FGY217 strain, which did not artificially express Ptr2p (Supplementary Fig. S1). The F-CUp assay system combined with growth analysis can also be a useful tool for developing an efficient fermentation medium.
Analysis using dipeptides (a) comprising histidine and leucine (b) comprising glycine and leucine. Pre-cultures of the BY4742 strain were diluted to give OD660=0.06 in each test medium (10 mM of each dipeptide, 0.14% yeast synthetic drop-out medium without leucine, 0.67% yeast nitrogen base without amino acids and 2% galactose) and incubated at 30 °C.
F-CUp assay using a dipeptide library
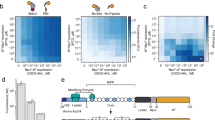
We performed a comprehensive analysis using a dipeptide library by the F-CUp assay system to characterize the substrate multispecificity of Ptr2p (Fig. 4a). For 338 types of dipeptides that could be synthesized, we calculated the Ki values of 237. Ki values could not be calculated for substrates for which the IC50 values were above 1.0 mM due to their poor solubility. Instead, their Ki values were assigned to be greater than 0.77 mM.
(a) Comprehensive analysis using a dipeptide library by the F-CUp assay system. Colours of cells correspond to Ki values. N.T.: not tested. Data are presented as means±s.d. (n=3). (b) Distribution of Ki values. (c) Frequency analysis of amino-acid residues with high or low-affinity dipeptides using the WebLogo programme ( http://weblogo.berkeley.edu/). Blue, red and pink arrowheads indicate N-end rule amino acids in yeast, essential amino acids and semi-essential amino acids for humans, respectively.
The calculated Ki values showed a wide range distribution (Fig. 4b). Trp-Phe exhibited the highest affinity (Ki=0.020 mM). It was also found that their affinities for Ptr2p were 2,400 times higher than those with the lowest affinity: Gly–Gly–Gly with Ki=48 mM analysed in this study. We selected a group of high-affinity dipeptides with Ki values below 0.077 mM and a group of low-affinity dipeptides with Ki values above 0.77 mM. We used these in an appearance frequency analysis of the amino-acid residues using the WebLogo programme (Fig. 4c). This showed that dipeptides containing aromatic amino acids (namely Phe, Trp and Tyr) and branched-chain amino acids (namely Ile, Leu and Val) frequently appeared in the high-affinity group. On comparing, it was found that the low-affinity group had a high frequency of negatively charged amino acids (that is, Asp and Glu), as well as amino acids that were predicted to influence peptide bond conformation (that is, Gly and Pro). In both groups of dipeptides, amino-acid residues at the amino terminus showed a higher propensity compared with those at the C-termini, which suggested that an amino-acid residue at the N-terminus had a more significant role in recognition by Ptr2p than those in the C-terminus.
Constructing ligand affinity prediction models
To expand the applications of our assay data, we constructed discrimination analysis models to predict ligand affinity in silico (Table 1). Our assay data comprised discrete over-threshold data (Ki>0.77) for several low-affinity dipeptides within continuous Ki data, along with future screening applications for in silico pre-screening; therefore, we selected discrimination analysis models to predict categorical labels for ligand molecules. Compared with conventional ligand prediction models, we selected features that could be simply calculated from the primary sequences of dipeptides as descriptive parameters to construct simple prediction models that utilizes fewer parameters.
We examined a total of six prediction models and observed that categorizing dipeptide samples as high, medium or low can predict affinities with extremely high accuracy (average prediction>84%). By comparing the data set type for whether or not it included intermediate Ki ligands, we observed that 97% prediction accuracy (data set type B, prediction model type M2) can be achieved for objectively screening ligands that would interact with Ptr2p. Even with data set type A modelled by M2, predictions were accurate in the area of ‘extremely high-affinity samples’, which indicated sufficient applicability for ligand screening (Supplementary Fig. S2).
By comparing these prediction model types, we observed that the combined information on amino-acid residues and chemical property information was most effective for obtaining an accurate prediction model. Prediction accuracy slightly increased by adding chemical property parameters that described total ligand molecular properties. However, sufficient prediction accuracy was achieved by converting the primary sequences of dipeptides into a few amino-acid indices. In addition, we observed that even without amino-acid information, total molecular chemical property information could be used as an alternative parameter to maintain similar prediction accuracy.
From the parameter selection process of constructing six discrimination analysis models, the manner of a dipeptide–Ptr2p interaction could be determined. During the modelling process with model M1, index 3 (side-chain contribution to protein stability) at the N-terminal, index 14 (side-chain interaction parameter) and indices 8 and 1 (isoelectric point) were the first four parameters that greatly contributed to the increased prediction accuracy. These results were common physicochemical rules for interactions between dipeptides and Ptr2p, which could be extracted from our comprehensive dipeptide library assay data. This extraction rule by model analysis was only possible with the affinity data with variety, and would not be attainable from partial positive screening data obtained from conventional, limited size assays. In addition to the discriminant analysis models that predicted the categories of ligand affinities, multiple regression models that predicted Ki values directly from molecular properties were observed to provide sufficient accuracy for screening (corrected R2 values>0.734).
PTR2 gene expression controlled by an N-end rule dipeptide
To examine the effect of amino-acid sequence of dipeptide on PTR2 gene expression, we analysed yeasts grown in YPD media containing dipeptides Ala–Ala or Trp–Ala (Fig. 5). In a direct analysis of gene expression by the FGY217 strain using real time RT–PCR, PTR2 gene expression increased twofold by adding the N-end rule dipeptide Trp–Ala in the YPD medium as compared with adding the non N-end rule dipeptide Ala–Ala.
Discussion
In this study, the substrate multispecificity of Ptr2p, the major peptide transporter of S. cerevisiae, was characterized using an F-CUp assay system (Fig. 4). Although there have been several reports regarding affinity analyses of substrates for POT family members7,27,28,29, this is the first study to use a dipeptide library for a comprehensive analysis. From this library assay data, we also successfully constructed in silico ligand affinity prediction models for Ptr2p using discriminant analysis models. By analysing the model construction processes, new insights were obtained to further understand the manner of interactions between dipeptides and Ptr2p.
The primary structure of Ptr2p was compared with that of other family members to gain a better understanding of the substrate recognition mechanism of a POT family protein. Although the similarity between the primary structure of Ptr2p and that of PepTso was 33.7% (Supplementary Fig. S3), three dimensional structures suggested that 12 amino-acid residues comprising the substrate-binding site of PepTso were highly conserved among these family members (Fig. 6a). For Ptr2p, 11 amino-acid residues, excluding Ser100 (corresponds to Arg32 in PepTso)(Supplementary Fig. S3), were functionally similar to those of PepTso. Dipeptides that comprise aromatic amino acids displayed high affinity for Ptr2p, whereas dipeptides that comprise negatively charged amino acids displayed low affinity for Ptr2p (Fig. 4c). The results obtained using individual substrates indicated that the substrate preferences of hPEPT1, hPEPT2, PepTst and YjdL were similar to those of Ptr2p (Fig. 6b)7,15,30. This consistency likely indicates that POT family members share a common substrate recognition mechanism. This was supported by the fact that the substrate-binding sites of these family members comprise highly conserved amino-acid residues.
(a) Close view of the substrate-binding site structures. Homology models for Ptr2p and hPEPT1 were constructed using the SWISS-MODEL programme ( http://swissmodel.expasy.org/) with PepTso (PDB: 2XUT) as a template14. Structures of Ptr2p and hPEPT1 are superimposed on the PepTso structure. The PepTso structure is shown as a ribbon model and the amino-acid residues that interact with a substrate peptide in PepTso are shown as a stick model. Blue: PepTso, yellow: Ptr2p, red: hPEPT1. Putative binding substrates are shown as pink and blue circles. (b) Dipeptide affinity for each transporter. Ki values for hPEPT1 and hPEPT2 are from published data7. ND: not determined (Ki>0.77 mM). Yellow column: Ptr2p, red column: hPEPT1, blue column: hPEPT2.
In S. cerevisiae, Ptr2p expression is regulated by the N-end rule pathway31,32,33, by which the binding of dipeptides with a certain N-terminus to Ubr1p promotes the degradation of Cup9p, which is a repressor of the PTR2 gene. Thus, the Ptr2p expression level increases when peptides that meet the N-end rule are imported. Cai et al.22 reported that several N-end rule peptides were preferentially recognized by Ptr2p. This is consistent with our present results obtained from a comprehensive analysis of a dipeptide library. The N-terminus of our high-affinity group is consistent with amino acids that adhered to the N-end rule (Trp, Phe, His, Tyr, Leu, Ile and Lys; Fig. 4c). Interestingly, most of the essential and semi-essential amino acids for humans (Trp, Phe, His, Tyr, Met, Leu, Ile, Val, Lys and Arg) were also the constituent amino acids of high-affinity peptides (Fig. 4c). In general, the biosynthesis of aromatic amino acids requires the expression of multiple enzymes and involves energy-consuming reactions, whereas biosynthesis of acidic amino acids occurs from shorter branches of the TCA cycle34. The substrate multispecificity of the POT family of transporter, revealed by analysing Ptr2p, indicates that these family members are involved in the preferential uptake of specific amino acids that impose a biosynthesis burden on organisms (Fig. 4c). Based on this perspective, we propose the following positive feedback model for peptide uptake into S. cerevisiae (Fig. 7). (i) Peptides with vital amino acids are preferentially transported into S. cerevisiae cells based on the substrate multispecificity of Ptr2p. (ii) Peptide uptake is accurately sensed by the N-end rule pathway via Ubr1p. (iii) Subsequently, Ptr2p expression is promoted after Cup9p degradation. (iv) S. cerevisiae cells can then more efficiently absorb those vital amino acids in their peptide form. Enhanced PTR2 gene expression by a transported dipeptide was experimentally demonstrated using an N-end rule dipeptide, Trp–Ala (Fig. 5). The association between the substrate multispecificity of Ptr2p and the regulatory system for PTR2 gene expression is biologically reasonable.
(a) PTR2 gene expression is repressed by Cup9p. (b) Peptides with vital amino acids are preferentially transported into yeast cells based on the substrate multispecificity of Ptr2p. (c) Peptide uptake is accurately sensed by the N-end rule pathway via Ubr1p, following which Ptr2p expression is further promoted after Cup9p degradation31,32,33. (d) S. cerevisiae cells can then more efficiently absorb these vital amino acids in their peptide form.
By examining our prediction models, we observed that a few simple parameters that could be obtained from ligand sequence information could produce high accuracy ligand prediction models. The accuracy and the construction processes indicated that a few physicochemical properties of dipeptides were sufficient for discriminating their affinities for Ptr2p. Our prediction model accuracy strongly suggests that Ptr2p recognition is primarily governed by ‘property combinations’ that characterize the physiochemical properties of ligands rather than their exact sequence motifs. By comparing different types of descriptive parameters that can be derived from the same dipeptide sequence (M1, M2 and M3 comparisons in Table 1), we observed that the best prediction accuracy can be obtained when both ‘amino-acid-specific physicochemical properties’ and ‘total molecular chemical property’ were used as descriptors for dipeptide molecules. However, similar affinity prediction accuracy for Ptr2p was retained even after eliminating the ‘amino acid-specific physicochemical properties’. These results indicate that our affinity prediction models constructed in silico are applicable to pre-screening for medical applications. To examine this possibility, we compared the experimental and predicted binding affinities to Ptr2p for six medical compounds as a trial (Supplementary Table S1). The predicted results matched F-CUp assay results for alafosfalin, arphamenine B, valacyclovir and captopril, although they did not match for fosinopril and benazepril. Based on the distribution of their molecular sizes, we assumed that the constructed prediction model using dipeptide library affinity data could predict the affinities for molecules that were close in size to those of dipeptides, having an average molecular weight of 256. In other words, it was reasonable that the predicted performance was limited to the variations of the ‘molecular chemical properties’ that existed in the dipeptide library used for model training. Therefore, increasing the ‘molecular information’ to train for wider variations in transporter affinities along with adding descriptive parameters will be the focus of our next investigation to expand our prediction approach. However, despite its limited application, it should be noted that our F-CUp assay-derived exhaustive molecular interaction data together with this modelling concept has great potential for affinity predictions of dipeptide-like molecules with high accuracy. Compared to the transported prediction approaches demonstrated by Biegel et al.25, our affinity prediction approach is sufficient for affinity predictions using only simple and feasibly calculated parameters for molecules without any prior ligand structural information.
The model construction process also guided our understanding of dipeptide interactions with Ptr2p. In M1 models (Table 1), index 3 (side-chain contribution to protein stability) at the N-terminal, index 14 (side-chain interaction parameter) and indices 8 and 1 (isoelectric point) were reproducibly selected in the same order in two models using data set type A or B. Index 3 and index 14 are strongly related to residues with ‘aromatic rings’ and ‘interactive residues’ that can stabilize molecular interactions (Supplementary Table S2). Indices 8 and 1 and some parts of index 14 can be interpreted as ‘effect of charged and polarized residues’. Therefore, these two physicochemical property features can be considered as the main determining factors for Ptr2p ligand recognition. Several substrate characteristics for binding have been elucidated based on the interaction data for many ligands with POT family transporters. For PEPT1, binding models were proposed by Foley et al., Brandsch et al., and Daniel and Kottra23,35,36. According to these models, the properties of the individual residues at both the N- and C-termini are important. While the interactions between a peptide backbone, side-chains and the binding pocket are not completely understood, all of these models indicate that a bulky, hydrophobic side chain is advantageous for high affinity towards PEPT1. On comparing, acidic amino acids in the N-terminus resulted in a greater reduction in affinity than did the same amino acids in the C-terminus. These models for PEPT1 are consistent with our results based on an in silico analysis of Ptr2p in nature. Therefore, from a general point of view, this also supports the hypothesis that POT family members share a common substrate recognition mechanism.
Most organisms, including yeasts and humans, have both peptide transporters and amino-acid transporters and both types of transporters cooperatively contribute to amino-acid resource uptake. The rates of substrate uptake by peptide transporters are higher than those of amino-acid transporters4,5,6. Therefore, an important role of peptide transporters is to import amino acids that are found in bulk in the extracellular fluid with high efficiency. To perform this role, peptide transporters must have substrate multispecificity in order to recognize a variety of compounds. Ligand recognition by hydrophobic interactions as well as by π−π bonds, which are not strictly directional, is suitable for this purpose. It is likely that POT family proteins have evolved so that these transporters have become equipped with such a substrate recognition mechanism (Table 1). The fact that many of the amino-acid residues that are involved in substrate binding are aromatic amino acids supports these characteristics (Fig. 6)14,15. An ‘ambiguous’ substrate recognition mechanism, which is primarily based on physicochemical properties with no strict directionality, is the basis for substrate multispecificity and causes POT family members to act as drug transporters to absorb drugs, which are not natural substrates37,38,39. This ‘ambiguous’ substrate recognition mechanism was also observed in our regression analysis. The concept of affinity prediction for POT family members using combinations of amino-acid indices has a great potential to be extended to other targets for predicting the affinities of as many as 8,000 tripeptides40. The concept of using combinations of physicochemical properties for affinity prediction can also be applied to the information of structural properties used in structure-based drug design, which is becoming possible owing to the elucidation of the crystal structures of POT family members14,15. Therefore, the analytical data from this research provides important information for detailed pharmacophore mapping.
Methods
Materials and chemicals
β-Ala–Lys (AMCA) was purchased from Biotrend (Cologne, Germany). Substrate dipeptides were purchased from Anaspec (California, USA). The S. cerevisiae strains BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) and BY4742-ptr2Δ were purchased from Open Biosystems (Alabama, USA).
Preparation of Ptr2p-expressing cells (SC-Ptr2p)
PCR was used to isolate the PTR2 gene from the genome of S. cerevisiae FGY217 (MATa, ura3-52, lys2Δ201, pep4Δ). The gene-specific primers 5′-ACCCCGGATTCTAGAACTAGTGGATCCCCC ATGCTCAACCATCCCAGCCAAG-3′ and 5′-AAATTGACCTTGAAAATATAAATTTTCCCCTCACTTGTCATCGTCGTCCTTGTAGTCATATTTGGTGGTGGATCTTAGAC-3′ were used to obtain PCR fragments of the PTR2 gene. These primers contained gene-specific regions (bold) and homologous regions (Italic). A PCR fragment and the SmaI-linearized pRS426 GAL1 vector were co-transformed into BY4742-ptr2Δ cells. These two DNAs were then linked via homologous recombination41,42,43. Cells were spread on a selection plate (2% agar, 0.2% yeast synthetic drop-out medium without uracil, 0.67% yeast nitrogen base without amino acids and 2% glucose). Transformants were cultured at 30 °C for 48 h. The resulting transformants were Ptr2p-expressing cells: SC-Ptr2p. For the expression of Ptr2p, SC-Ptr2p cells were grown in selection medium (0.2% yeast synthetic drop-out medium without uracil, 0.67% yeast nitrogen base without amino acids and 2% glucose). After pre-culture at 30 °C for 24 h, cells were diluted to give an OD660=0.06 in induction medium (0.2% yeast synthetic drop-out medium without uracil, 0.67% yeast nitrogen base without amino acids and 2% galactose) cultured at 30 °C for 24 h.
Confocal microscopy
To express the Ptr2p–GFP fusion protein, 5′-AAATTGACCTTGAAAATATAAATTTTCCCC ATATTTGGTGGTGGATCTTAGAC-3′ was used as a reverse primer. Expression of the Ptr2p–GFP fusion protein used the same method as for Ptr2p. The localization of the Ptr2p–GFP fusion protein was analysed by detecting GFP fluorescence using an LSM-700 (Carl Zeiss MicroImaging, New York, USA).
Spot assay
SC-Ptr2p cells were grown in pre-culture medium at 30 °C for 24 h. Cells were spotted on an assay plate (10 mM His–Leu dipeptide, 0.14% yeast synthetic dropout medium without histidine, leucine and uracil, 0.67% yeast nitrogen base without amino acids, 2% galactose and 2% agar) and cultured at 30 °C for 5 days.
Fluorescence-based Competitive Uptake (F-CUp) assay
After induction, SC-Ptr2p cells were harvested by centrifugation and re-suspended in F-CUp assay buffer (150 mM NaCl, 50 mM Na-phosphate buffer; pH 6.0). Cells were harvested by centrifugation and re-suspended in F-CUp assay buffer to OD660=15. Cells were incubated with 0.05 mM β-Ala–Lys (AMCA) as a tracer substrate and at arbitrary concentration for oligopeptide analysis at 30 °C for 60 min. β-Ala–Lys (AMCA) is a dipeptide containing a fluorophore. Cells were washed three times with the F-CUp assay buffer. β-Ala–Lys (AMCA) uptake was quantified by whole-cell fluorescence (excitation at 355 nm and emission at 460 nm) using Flexstation III (Molecular Devices, California, USA). Km values were estimated using Lineweaver–Burk plot. The initial velocity of a reaction was determined from the fluorescence intensity increase during the first hour. At this point, the substrate concentration remained the same as the initial condition. The IC50 value was estimated based on the reduced fluorescence by competitive inhibition of the tracer uptake.
The Ki value was calculated using the Cheng–Prusoff equation from the Km of β-Ala–Lys (AMCA) and the IC50 of the analysed oligopeptide26. The mean Km value was used to calculate Ki values. Thus, the error for estimating Km values was not included in the error when determining Ki. This competition assay system can estimate the affinity of a substrate for a peptide transporter. Some dipeptides were actually transported by yeast cells based on their Ki values for Ptr2p (Fig. 3), although affinity does not always mean that a compound will be transported.
Cell growth analysis
For time course analysis of culture turbidity, strain BY4742 was grown overnight in culture medium (0.2% yeast synthetic medium, 0.67% yeast nitrogen base without amino acids and 2% glucose) at 30 °C for 24 h. Pre-cultures were diluted to give OD660=0.06 in each test medium (10 mM each dipeptide, 0.14% yeast synthetic dropout medium without leucine, 0.67% yeast nitrogen base without amino acids and 2% glucose) and incubated at 30 °C. Culture turbidity was monitored by measuring OD660 using a Biophotorecorder TVS 062CA (Advantec, Tokyo, Japan).
Ligand affinity prediction models and discrimination analysis
Using the data set of dipeptide sequences in conjunction with their affinity data from the F-CUp assay, dipeptide affinity prediction models were constructed using discrimination analysis with PASW version 18, release 18.0.0 (IBM Corporation, Armonk, NY, USA). For objective variables (that is, teaching signals), experimentally determined Ki values were grouped into three categories: low Ki samples with Ki<0.1, N=102; high Ki samples with Ki>0.77, N=100; and medium Ki for the remaining samples, N=135.
For predictor variables (that is, input parameters), dipeptide sequences were converted into two types of parameters. Type I parameters included seven amino-acid indices, which were calculated by converting each amino acid at either the N-terminal or the C-terminal side of dipeptides using amino-acid indices (AA index1, Genome Net Japan, organized by Kyoto University; http://www.genome.jp/dbget-bin/www_bfind?aaindex1)44. All indices in the database (N=544, version 9.1, as of August 2006) were previously analysed by hierarchical clustering; seven indices45,46,47,48,49,50 were chosen as independent indices that represented seven major independent clusters, which demonstrated no multicolinearity (Supplementary Table S2). Type II parameters included 10 physicochemical property parameters, which were obtained from each total dipeptide molecule with an original pipeline protocol using Chemistry Component Collection in Pipeline Pilot (Accelrys, San Diego, CA, USA).
For modelling, two different prediction concepts were compared with two types of data sets: type A data sets to construct prediction models to discriminate all three types of Ki samples (high, medium and low); and type B data sets to construct prediction models to discriminate either low Ki samples or high Ki samples. For both prediction concepts, three different prediction models were constructed: M1 prediction models using only type I parameters; M2 prediction models using type I and type II parameters and M3 prediction models using only type II parameters for comparing model performances between the different types of parameters. During the model construction process, a parameter increasing and decreasing method was employed with a threshold of P<0.20 based on F-Test for parameter selection. Model accuracy was evaluated based on the discrimination accuracy of either each teaching signal category or as a total.
Real time RT–PCR
A FGY217 strain pre-culture was diluted to give OD660=0.06 in each test YPD medium (2% peptone, 1% yeast extract, 2% glucose and 10 mM Trp–Ala or Ala–Ala), and then grown at 30 °C for 5 h. Yeast total RNA was isolated using a NucleoSpin RNA II kit (Machery-Nagel, Diiren, Germany) according to the manufacturer's protocol. The amount of total RNA was quantified by monitoring absorbance at 260 nm. The first strand was synthesized using a PrimeScript RT reagent kit (Takara, Shiga, Japan). Quantitative real time PCR analysis was done with a Thermal Cycler Dice Real Time System (Takara, Shiga, Japan) using SYBR Premix EX Taq (Takara, Shiga, Japan) and specific primers. The following gene-specific primers were used: 5′-CCACCATGTTCCCAGGTATT-3′ and 5′-CCAATCCAGACGGAGTACTT-3′ for ACT1; and 5′-CAGTGACCGTTGATCCTAAAT-3′ and 5′-CTGAAGCACAACCAGAACAAA-3′ for PTR2. PTR2 mRNA levels were normalized to those of ACT1 values using the 2-ΔΔCT method. We calculated the fold-change of PTR2 mRNA in YPD medium containing Trp–Ala compared to that containing Ala–Ala.
Additional information
How to cite this article: Ito, K. et al. Analysing the substrate multispecificity of a proton-coupled oligopeptide transporter using a dipeptide library. Nat. Commun. 4:2502 doi: 10.1038/ncomms3502 (2013).
References
Daniel, H., Spanier, B., Kottra, G. & Weitz, D. From bacteria to man: archaic proton-dependent peptide transporters at work. Physiology (Bethesda) 21, 93–102 (2006).
Gomolplitinant, K. M. & Saier, M. H. Jr. Evolution of the oligopeptide transporter family. J. Membr. Biol. 240, 89–110 (2011).
Steiner, H. Y., Naider, F. & Becker, J. M. The PTR family: a new group of peptide transporters. Mol. Microbiol. 16, 825–834 (1995).
Maebuchi, M. et al. Improvement in the intestinal absorpton of soy protein by enzymatic digestion to oligopeptide in healthy adult men. Food Sci. Technol. Res 13, 45–53 (2007).
Matthews, D. M. Intestinal absorption of peptides. Physiol. Rev. 55, 537–608 (1975).
Ito, K. et al. Soy peptides enhance heterologous membrane protein productivity during the exponential growth phase of Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 76, 628–631 (2012).
Biegel, A. et al. The renal type H+/peptide symporter PEPT2: structure-affinity relationships. Amino Acids 31, 137–156 (2006).
Fei, Y. J. et al. Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 368, 563–566 (1994).
Liang, R. et al. Human intestinal H+/peptide cotransporter. Cloning, functional expression, and chromosomal localization. J. Biol. Chem. 270, 6456–6463 (1995).
Adibi, S. A. The oligopeptide transporter (Pept-1) in human intestine: biology and function. Gastroenterology 113, 332–340 (1997).
Liu, W. et al. Molecular cloning of PEPT 2, a new member of the H+/peptide cotransporter family, from human kidney. Biochim. Biophys. Acta. 1235, 461–466 (1995).
Boll, M. et al. Expression cloning and functional characterization of the kidney cortex high-affinity proton-coupled peptide transporter. Proc. Natl Acad. Sci. USA 93, 284–289 (1996).
Shen, H. et al. Localization of PEPT1 and PEPT2 proton-coupled oligopeptide transporter mRNA and protein in rat kidney. Am. J. Physiol. 276, F658–F665 (1999).
Newstead, S. et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 30, 417–426 (2011).
Solcan, N. et al. Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J. 31, 3411–3421 (2012).
Rubio-Aliaga, I. & Daniel, H. Mammalian peptide transporters as targets for drug delivery. Trends Pharmacol. Sci. 23, 434–440 (2002).
Homann, O. R., Cai, H., Becker, J. M. & Lindquist, S. L. Harnessing natural diversity to probe metabolic pathways. PLoS Genet. 1, e80 (2005).
Cai, H., Kauffman, S., Naider, F. & Becker, J. M. Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics 172, 1459–1476 (2006).
Perry, J. R., Basrai, M. A., Steiner, H. Y., Naider, F. & Becker, J. M. Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol. Cell Biol. 14, 104–115 (1994).
Hauser, M., Narita, V., Donhardt, A. M., Naider, F. & Becker, J. M. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 18, 105–112 (2001).
Kitagawa, S. et al. Effect of soy peptide on brewing beer. J. Biosci. Bioeng. 105, 360–366 (2008).
Cai, H., Hauser, M., Naider, F. & Becker, J. M. Differential regulation and substrate preference in two peptide transporters of Saccharomyces cerevisiae. Eukaryot. Cell 6, 1805–1813 (2007).
Daniel, H. & Kottra, G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers. Arch. 447, 610–618 (2004).
Thesis, S. et al. Synthesis and characterization of high affinity inhibitors of the H+/peptide transporter PEPT2. J. Biol. Chem 277, 7287–7292 (2002).
Biegel, A., Gebauer, S., Brandsch, M., Neubert, K. & Thondorf, I. Structural requirements for the substrates of the H+/peptide cotransporter PEPT2 determined by three-dimensional quantitative structure-activity relationship analysis. J. Med. Chem 49, 4286–4296 (2006).
Cheng, Y. & Prusoff, W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973).
Nour-Eldin, H. H. et al. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534 (2012).
Foltz, M., Meyer, A., Theis, S., Demuth, H. U. & Daniel, H. A rapid in vitro screening for delivery of peptide-derived peptidase inhibitors as potential drug candidates via epithelial peptide transporters. J. Pharmacol. Exp. Ther. 310, 695–702 (2004).
Geissler, S., Zwarg, M., Knütter, I., Markwardt, F. & Brandsch, M. The bioactive dipeptide anserine is transported by human proton-coupled peptide transporters. FEBS J. 277, 790–795 (2010).
Jensen, J. M. et al. Biophysical characterization of the proton-coupled oligopeptide transporter YjdL. Peptides 38, 89–93 (2012).
Byrd, C., Turner, G. C. & Varshavsky, A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 17, 269–277 (1998).
Turner, G. C., Du, F. & Varshavsky, A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405, 579–583 (2000).
Hwang, C. S. & Varshavsky, A. Regulation of peptide import through phosphorylation of Ubr1, the ubiquitin ligase of the N-end rule pathway. Proc. Natl Acad. Sci. USA 105, 19188–19193 (2008).
Braus, G. H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 55, 349–370 (1991).
Foley, D. W., Rajamanickam, J., Bailey, P. D. & Meredith, D. Bioavailability through PepT1: the role of computer modelling in intelligent drug design. Curr. Comput. Aided Drug Des. 6, 68–78 (2010).
Brandsch, M., Knütter, I. & Bosse-Doenecke, E. Pharmaceutical and pharmacological importance of peptide transporters. J. Pharm. Pharmacol. 60, 543–585 (2008).
Terada, T. & Inui, K. Peptide transporters: structure, function, regulation and application for drug delivery. Curr. Drug Metab. 5, 85–94 (2004).
Zhang, L. et al. Synthesis and evaluation of a dipeptide-drug conjugate library as substrates for PEPT1. ACS Comb. Sci 14, 108–114 (2012).
Kouodom, M. N. et al. Toward the selective delivery of chemotherapeutics into tumor cells by targeting peptide transporters: tailored gold-based anticancer peptidomimetics. J. Med. Chem 55, 2212–2226 (2012).
Kaga, C., Okochi, M., Tomita, Y., Kato, R. & Honda, H. Computationally assisted screening and design of cell-interactive peptides by a cell-based assay using peptide arrays and a fuzzy neural network algorithm. Biotechniques 44, 393–402 (2008).
Ito, K. et al. Advanced method for high-throughput expression of mutated eukaryotic membrane proteins in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 371, 841–845 (2008).
Drew, D. et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 3, 784–798 (2008).
Newstead, S., Kim, H., von Heijne, G., Iwata, S. & Drew, D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 104, 13936–13941 (2007).
Kawashima, S. et al. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 36, D202–D205 (2008).
Zimmerman, J. M., Eliezer, N. & Simha, R. The characterization of amino acid sequences in proteins by statistical methods. J. Theor. Biol. 21, 170–201 (1968).
Fauchere, J. L., Charton, M., Kier, L. B., Verloop, A. & Pliska, V. Amino acid side chain parameters for correlation studies in biology and pharmacology. Int. J. Pept. Protein Res. 32, 269–278 (1988).
Takano, K. & Yutani, K. A new scale for side-chain contribution to protein stability based on the empirical stability analysis of mutant proteins. Protein. Eng. 14, 525–528 (2001).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Crawford, J. L., Lipscomb, W. N. & Schellman, C. G. The reverse turn as a polypeptide conformation in globular proteins. Proc. Natl Acad. Sci. USA 70, 538–542 (1973).
Krigbaum, W. R. & Komoriya, A. Local interactions as a structure determinant for protein molecules: II. Biochim. Biophys. Acta 576, 204–248 (1979).
Acknowledgements
This study was supported in part by a Research Grant from the Society for Research on Umami Taste, a Grand-in-Aid for Young Scientists (B) (2370139) and the Fuji Foundation for Protein Research. The authors would like to thank Enago ( www.enago.jp) for the English language review.
Author information
Authors and Affiliations
Contributions
K.I. established the F-CUp assay system, designed the project, organized the entire research and wrote the manuscript. A.H. performed the exhaustive analysis of dipeptide library analysis using the F-CUp assay system and cell growth analysis. V.T.T.L. and S.K. (Sayuri Kitagawa) established the F-CUp assay system. S.K. (Shun Kawai) and T.M. performed the discrimination analysis. Y.Y. performed the RT–PCR analysis. R.K. supervised the discrimination analysis and wrote the section of in silico analysis. Y.K. wrote the manuscript and co-organized the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S3 and Supplementary Tables S1-S2 (PDF 1291 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Ito, K., Hikida, A., Kawai, S. et al. Analysing the substrate multispecificity of a proton-coupled oligopeptide transporter using a dipeptide library. Nat Commun 4, 2502 (2013). https://doi.org/10.1038/ncomms3502
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms3502
This article is cited by
-
Genome-wide analysis of PTR transporters in Candida species and their functional characterization in Candida auris
Applied Microbiology and Biotechnology (2022)
-
Enhancement of protein thermostability by three consecutive mutations using loop-walking method and machine learning
Scientific Reports (2021)
-
Intestinal membrane transporter-mediated approaches to improve oral drug delivery
Journal of Pharmaceutical Investigation (2021)
-
Global mapping of protein–metabolite interactions in Saccharomyces cerevisiae reveals that Ser-Leu dipeptide regulates phosphoglycerate kinase activity
Communications Biology (2021)
-
Tracking dipeptides at work-uptake and intracellular fate in CHO culture
AMB Express (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.