Abstract
Phosphorus supply is a major factor responsible for reduced crop yields. As a result, plants utilize various adaptive mechanisms against phosphorus depletion, including lipid remodelling. Here we report the involvement of a novel plant lipid, glucuronosyldiacylglycerol, against phosphorus depletion. Lipidomic analysis of Arabidopsis plants cultured in phosphorus-depleted conditions revealed inducible accumulation of glucuronosyldiacylglycerol. Investigation using a series of sulfolipid sulfoquinovosyldiacylglycerol synthesis-deficient mutants of Arabidopsis determined that the biosynthesis of glucuronosyldiacylglycerol shares the pathway of sulfoquinovosyldiacylglycerol synthesis in chloroplasts. Under phosphorus-depleted conditions, the Arabidopsis sqd2 mutant, which does not accumulate either sulfoquinovosyldiacylglycerol or glucuronosyldiacylglycerol, was the most severely damaged of three sulfoquinovosyldiacylglycerol-deficient mutants. As glucuronosyldiacylglycerol is still present in the other two mutants, this result indicates that glucuronosyldiacylglycerol has a role in the protection of plants against phosphorus limitation stress. Glucuronosyldiacylglycerol was also found in rice, and its concentration increased significantly following phosphorus limitation, suggesting a shared physiological significance of this novel lipid against phosphorus depletion in plants.
Similar content being viewed by others
Introduction
Phosphorus (P) is an essential component in cells and has a crucial role in cellular functionality and metabolism. However, because of limited P availability in many soils, plants often suffer from P deficiency1. Indeed, it is estimated that crop yields for 40% of the world’s arable land are reduced because of P unavailability2. As current known P reserves are limited and P-based fertilizers are economically produced, efficient use and the minimization of the loss of P resources (that is, recycling of struvite, magnesium ammonium phosphate) are very important to sustainable agriculture3,4,5. However, plants have developed several adaptation mechanisms to cope with P-limited environments.
P mobilization by membrane lipid remodelling, that is, changing the composition of the membrane lipids rich in phospholipids, is one of the general mechanisms of adapting to P limitation in plants6,7,8,9. Replacement of membrane phospholipids with non-phosphorus glycerolipids such as sulfoquinovosyldiacylglycerol (SQDG) and digalactosyldiacylglycerol (DGDG), which promote the remobilization of P, is typical of metabolic signatures associated with lipid remodelling during phosphate deprivation. These glycolipids are usually found in chloroplast membranes, but DGDG is also found in extraplastidic membranes in P-starved plants7,8,10. Thus far, various aspects of lipid remodelling following P limitation have been investigated. The biosynthetic enzymes involved in the enhanced production of these non-phosphorus glycolipids under P limitation, mainly glycosyltransferases and sugar-donor synthetic enzymes, have been intensely characterized11,12,13,14,15,16,17,18. Phospholipid degradation and P recycling have also been analysed through the characterization of several phospholipases and glycerophosphodiesterases15,19,20,21,22. In addition, parts of the signal transduction pathway for lipid remodelling have been studied14,15,23,24. However, detailed metabolite analysis regarding lipid remodelling has been limited. Previous studies have thus far focused only on several well-known lipid classes25 because of the difficulties associated with the comprehensive analysis of plant lipids, which consist of a wide variety of hydrophobic chemicals. In this study, we applied untargeted metabolomic analysis, which allows comprehensive chemical metabolic phenotyping of cells, to investigate lipid remodelling in detail and to identify a new key lipid molecule that has an essential role in protection against P-depletion stress.
Results
A new plant lipid inducibly accumulates under P depletion
To expose plants to P-limiting stress, wild-type Arabidopsis thaliana plants grown on phosphate (Pi)-sufficient growth medium for 14 days were transferred to either Pi-sufficient or Pi-depleted growth medium. After another 14 days of growth, lipids were extracted from the leaves and subjected to lipidomic analysis by hydrophilic interaction chromatography (HILIC) coupled with ion trap time-of-flight mass spectrometry26. This analysis indicated that P limitation induced a decrease in phospholipids such as phosphatidylglycerol (PG), phosphatidylethanolamine (PE), phosphatidylinositol (PI) and phosphatidylcholine, with a complementary increase in non-phosphorus glycerolipids, including SQDG (~4.5-fold) and DGDG (~2-fold), as previously reported (Fig. 1a)7,12. The total monogalactosyldiacylglycerol (MGDG) level did not significantly change in response to P limitation, as described below (Fig. 1a). In addition to these changes, a new peak (UK1; retention time, 11.4 min) was found to appear under P limitation but not under P sufficiency. To obtain detailed information regarding the overall metabolic changes induced by P limitation, an orthogonal projection to a latent structure-discriminant analysis (OPLS-DA) model was examined using the lipidomic data obtained from the plants subjected to P-limited and P-sufficient conditions27. The OPLS-DA (Supplementary Fig. S1) and OPLS loading S-plot (Fig. 1b) revealed that the discriminative inducible metabolites associated with the P-limiting treatment were mainly UK1, SQDG and DGDG (Table 1), and the significantly decreased metabolites were phospholipids (Table 2). We focused on the new lipid UK1 for further study.
(a) Wild-type Arabidopsis plants grown under P-sufficient conditions were transferred to either P-sufficient (+P, blue line) or P-depleted (−P, red line) medium. After cultivation for 14 days, crude lipid extracts from the leaves were analysed by HILIC-MS. The total ion current detected by the negative ion mode is shown. An unknown lipid (denoted as UK1, further identified as GlcADG) appeared only under P-limited conditions. (b) S-plot of OPLS-DA based on ANOVA of the cross-validated residuals (CV-ANOVA). The S-plot demonstrates the covariance versus the correlation in conjunction with the variable trend plots and allows easier visualization of the data. The variables that showed maximum change, and therefore strongly contributed to the class separation, are plotted at the top or bottom of the S-shaped plot. The variables that did not significantly vary are plotted in the middle. Each point represents a lipid molecule detected in the negative ion mode. Red stars denote the [M–H]− ions of the UK1 species, indicating that the change in the levels of a series of UK1 molecules is highly associated with P-limiting treatment. The molecules at the top and bottom are listed in Tables 1 and 2.
Mass spectral analyses revealed that UK1 is a lipid class consisting of several similar molecular species whose molecular weights differed by 2 Da (Supplementary Fig. S2a,c). The tandem mass spectrum (MS/MS) of UK1 further revealed that the major product ions were attributable to two fatty acids and glucuronosylglycerol (Fig. 2a, and Supplementary Fig. S2b), which suggests that UK1 is a glucuronosylated diacylglycerol (Fig. 2b). MS/MS results in the positive ion mode, exhibiting the neutral loss attributable to glucuronic acid, also supported this conclusion (Supplementary Fig. S2c,d). The presence of 1,2-diacyl-3-O-α-glucuronosylglycerol (GlcADG, 1) has been reported in several bacteria and fungi and in two species of unicellular algae28,29,30, but it has not yet been described in higher plants. In addition, the physiological function and biosynthetic gene have not been identified in these organisms. Thus, we purified UK1 from A. thaliana and GlcADG from the proteobacterium Brevundimonas diminuta, previously called Pseudomonas diminuta31, and compared them with respect to several physicochemical properties.
(a) MS/MS of [M–H]− ion of the major UK1 molecule (34:3 UK1. m/z 765.517). FA, fatty acid. The asterisk indicates the key fragment ion attributable to the glucuronosylglycerol moiety. (b) Comparison of the chromatographic behaviour of UK1 and GlcADG isolated from B. diminuta. The structure of the most major GlcADG molecule from Arabidopsis (34:3 GlcADG) is presented. The species labels, namely, total acyl carbons:total acyl double bonds, are shown in parentheses. Fragment analysis in the negative mode allowed annotation of fatty acyl groups and tentative assignment of these groups to the sn-1 and sn-2 positions60.
The new plant lipid was identified as GlcADG
The purified UK1 and bacterial GlcADG showed the same chromatographic behaviour (Fig. 2b) and same fragmentation pattern (Fig. 2a, and Supplementary Figs S3,S4), supporting the hypothesis that UK1 was GlcADG. However, further characterization of the sugar moiety was still required to identify UK1. At first, we intended to analyse the NMR spectra of UK1 using bacterial GlcADG as the reference; however, NMR spectra of GlcADG from B. diminuta were not available because the bacterial GlcADG was identified via several other methods of chemical derivatization31. Thus, we carefully analysed the structure of these two lipids from two different organisms.
First, GlcADG from B. diminuta was hydrolysed with hydrochloric acid, and the hydrolysate was analysed using GC–MS. The compound found in the hydrolysate was identified as glucuronic acid, not galacturonic acid, by comparison with the commercially available standard (Supplementary Figs S5,S6), indicating that the glycosyl moiety of isolated bacterial lipid is glucuronic acid. Then, we analysed NMR spectra of the methyl ester of GlcADG from B. diminuta. The 1H and 13C NMR data on the methyl ester of bacterial GlcADG is very similar to previously published data on GlcADG from Caulobacter bacteroides32 and Aspergillus fumigates33, respectively, considering the effect of methylation of the carboxyl group of glucuronic acid. In addition, the 1H and 13C NMR spectra of the methyl ester of UK1 are also very similar to those of the methyl ester of bacterial GlcADG. Thus, the lipids isolated from A. thaliana and B. diminuta are both GlcADG (1). The coupling constants of the anomeric proton (H-1′) of GlcADG from A. thaliana and B. diminuta are both 3.7 Hz, indicating that the configuration of the glycosidic bond in these lipids is ‘alpha’. The coupling pattern of protons in the uronic acid moiety is also consistent with the conclusion that the sugar moiety of this lipid class is glucuronic acid.
Elucidation of the possible biosynthetic pathway of GlcADG
The biosynthesis of lipids occurs in the discrete subcellular biogenic membranes of different cellular compartments, and the fatty composition of the glycerolipids synthesized in one compartment is quite different from those synthesized in other compartments. For example, endoplasmic reticulum-derived glycerolipids have an 18-carbon fatty acid at sn-2 of glycerol, while glycerolipids synthesized de novo in plastids have a 16-carbon fatty acid at sn-2 (refs 34,35). Some of these lipids are transported from one compartment to another and transformed into different lipid classes by changing their polar headgroup. This transformation is catalysed by several different enzymes that are not evenly distributed in every organelle. These mechanisms for membrane lipid biosynthesis make the fatty acid composition and diacylglycerol (DAG) composition of each lipid class unique. Thus, the MS and MS/MS spectral information obtained as lipidomic data were further utilized to elucidate the biosynthetic mechanism of GlcADG production, based on the assumption that each glycerolipid class possesses a characteristic fatty acid composition that reflects their synthetic route. Hierarchical clustering (HCL) of the z-scored DAG composition of each lipid class demonstrated that the DAG composition of GlcADG and SQDG were nearly identical, indicating that GlcADG belongs to the same group as the SQDG produced during P limitation (Fig. 3a). Analysis of the negative MS/MS data on these lipids further confirmed the high similarities between the fatty acid compositions of GlcADG and SQDG (Supplementary Fig. S3). It should be noted that all plant GlcADG species with acyl groups consisting of 34 carbons have a palmitic acid moiety at the sn-2 position (Supplementary Fig. S3). Palmitic acid (16:0 fatty acid) is a minor acyl group both in galactolipids and at sn-2 of phospholipids, but it is the major group at sn-2 in SQDG (Supplementary Fig. S3)34,35. These results collectively suggest that the biosynthesis of GlcADG may share, at least in part, the biosynthetic pathway of SQDG.
(a) HCL of glycerolipid classes based on the z-scored DAG compositions of the fatty acid moieties. Key: P-sufficient (+P), P-depleted (−P). To test the rigour of the presented dendrogram, we performed a permutation test using jackknife resampling (permutation, 1,000). The colours of the branches indicate the level of branch rearrangement reliability (jackknife proportions) (black: 95–100% support, blue: 80–90% support). (b) Levels of individual lipid classes are expressed as relative abundance (Rel. ab.) against that of the wild-type grown under P-sufficient conditions. GlcADG was not detected under the P-sufficient condition. Each data point represents the mean±s.d. value of four experiments. Different letters indicate statistically significant differences (P<0.05, Tukey’s test) among the tested genotypes grown under identical growth conditions. The absence of letters demonstrates no significant difference among all the tested genotypes for a lipid class. The asterisk denotes ‘not detected’.
SQDG synthase is required for GlcADG synthesis
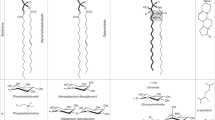
In A. thaliana, SQDG is synthesized in the chloroplast by three unique steps (Fig. 4a): synthesis of UDP-glucose (UDP-Glc) by UDP-Glc pyrophosphorylase 3 (UGP3)17, assembly of UDP-Glc and sulphite into UDP-sulfoquinovose (UDP-SQ) by UDP-SQ synthase (SQD1)11,36, and the subsequent transfer of the SQ moiety from UDP-SQ to DAG by SQDG synthase (SQD2)12,37. These three SQDG biosynthetic enzymes are located in the chloroplast, and the expression of their coding genes is coordinated in green tissues and significantly upregulated by P limitation11,12,17,37,38,39,40. Based on the above-mentioned hypothesis that GlcADG synthesis partly shares the pathway of SQDG synthesis, GlcADG accumulation was investigated in a series of mutants deficient in all three SQDG biosynthetic genes of A. thaliana under the P-depleted condition. The sqd1 and ugp3 mutants produced GlcADG at a slightly higher level than the wild-type control (Fig. 3b and Supplementary Fig. S7); strikingly, however, no accumulation of GlcADG was observed in the independent sqd2 mutants. These results clearly demonstrate that GlcADG is synthesized by the action of SQD2 (Fig. 4a), presumably using DAG and a sugar donor—most likely UDP-glucuronic acid (UDP-GlcA)—instead of UDP-SQ, which is used in the synthesis of SQDG. However, two other enzymes involved in SQDG synthesis, UGP3 and SQD1, are not involved in the synthesis of GlcADG.
(a) Three enzymes (UGP3, SQD1 and SQD2) are involved in the biosynthesis of SQDG in the chloroplasts of Arabidopsis. SQD2 is responsible for SQDG synthesis under P-sufficient conditions. Under P-deficient conditions, SQD2 produces not only SQDG but also GlcADG. (b) Wild-type plants and a series of SQDG-deficient mutants of A. thaliana grown under P-sufficient conditions were transferred to either P-sufficient or P-depleted medium. After cultivation for 4 weeks, the growth of each genotype was compared. The individuals showing the most typical phenotype of P deprivation are displayed. Only 2 sqd2 mutants appeared bleached under P-starvation conditions. Scale bar, 1 cm. Images of other individuals are shown in Supplementary Fig. S11.
GlcADG deficiency leads to significant growth defects
An Arabidopsis mutant with disrupted SQD2 demonstrates impaired growth in the P-limited condition12. Therefore, we carefully examined the lipid profiles and growth of a series of SQDG-deficient Arabidopsis mutants with identical genetic backgrounds. Under the P-sufficient condition, the total levels of PG were slightly higher in the SQDG-deficient mutants than in the wild-type (Fig. 3b), as previously reported11,12,17. Detailed analysis revealed that 34:2–34:4 PG and 34:3 PI accumulated more in the SQDG-deficient mutants than in the wild-type (Supplementary Fig. S8). Upon P limitation, the ugp3 and sqd1 mutants accumulated GlcADG at slightly higher levels than the wild-type (Fig. 3b and Supplementary Fig. S9). In addition, the decrease of PI, PG and PE following P limitation was suppressed in the SQDG-deficient mutants, as the total levels of GlcADG and SQDG were lower (Fig. 3b). In particular, 34:2–34:3 PI and 34:2–34:3 and 36:5 PE were significantly elevated in the sqd2 mutant compared with the other tested genotypes (Supplementary Fig. S9). In contrast to these significant changes in phospholipids, the deficiency of GlcADG and SQDG led to only a subtle effect on galactolipid (MGDG and DGDG) contents (Fig. 3b). The levels of glucosylceramide (GlcCer) and steryl glucoside (StGlc), which serve along with glycerolipids as membrane lipids, were also analysed. The amount of GlcCer and StGlc increased following P limitation, and the amounts of StGlc in P-starved sqd2 was higher compared with the other tested genotypes (Supplementary Fig. S10ab). There were no clear differences in the total levels of GlcCer among the tested genotypes when P starved, although there were some differences among their GlcCer profiles (Supplementary Fig. S10c). These results indicate that the lipid remodelling mechanisms induced by P limitation are significantly altered by GlcADG deficiency.
We then investigated the growth of the SQDG-deficient mutants under P-depleted conditions. Under P-sufficient conditions, all SQDG-deficient mutants and the wild-type demonstrated similar growth (Supplementary Fig. S11a). In contrast, under P-depleted conditions, the two independent sqd2 mutants that did not accumulate SQDG or GlcADG were severely damaged compared with the sqd1 and ugp3 mutants, in which SQDG is absent but GlcADG is still produced (Fig. 4b and Supplementary Fig. S11b). These results indicate that GlcADG has a major role in mitigating P-depletion stress in plants. In addition, the leaf colour of the P-starved sqd2 mutants appeared bleached. The total chlorophyll content was reduced in sqd2 mutants compared with wild-type, ugp3 or sqd1 when plants were grown under P depletion (Supplementary Fig. S12). These results indicate that GlcADG deficiency accelerates the breakdown of chlorophylls and the de-regulation of chloroplast functions.
As described above, the disruption of the SQD2 gene in Arabidopsis led to the de-regulation of lipid remodelling under P depletion, and the shoots of sqd2 mutants showed enhanced senescence compared with other SQDG-deficient mutants. These data suggest that Pi levels might be affected by disruption of SQD2. Thus, we analysed the inorganic Pi level in shoots grown under P-controlled conditions. The inorganic Pi levels in shoots were significantly altered as Pi availability changed, whereas there was no significant difference in the Pi content of the shoots of the tested genotypes (Supplementary Fig. S13). Thus, the severe growth defects observed in the shoots of sqd2 mutants under P-limited conditions are not necessarily related to the free Pi content in shoots, suggesting an association of GlcADG in the protection of the membrane environment under P-limited growth conditions.
To obtain further insight into the general role of GlcADG in P depletion in plants, the distribution of GlcADG was also investigated in rice (Oryza sativa L.). GlcADG was detected in the leaves of rice grown even under the nutrition-sufficient condition (Supplementary Fig. S14), and the levels were significantly elevated (5–7 times) by P deprivation, as observed in A. thaliana (Fig. 5). Significant lipid profile changes upon P limitation were also observed in rice leaves, as observed in A. thaliana (Supplementary Fig. S15). These results suggest a general physiological significance for this novel lipid under P-limited conditions across plant species.
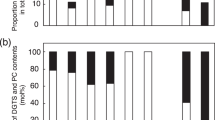
Comparison of GlcADG levels in the leaves of rice grown under either P-sufficient (+P) or P-depleted (−P) conditions. The levels of each lipid molecule are expressed as relative values compared with the level of 34:2 GlcADG in rice grown under P-sufficient growth conditions. Each data point represents the mean value of three experiments ±s.d. Asterisks indicate a statistically significant difference from the P-sufficient growth condition (*P<0.05, Welch’s t-test). More detailed data can be found in Supplementary Figs S14, S15.
Discussion
SQDG is an unusual glycerolipid because its polar headgroup consists of SQ, a 6-deoxy-6-sulfonate-glucose (Fig. 4). Much has been speculated about the possible function of this lipid in plants41. However, the isolation of SQDG-deficient bacterial mutants42,43, a mutant of the unicellular algae Chlamydomonas44, and a mutant of the plant A. thaliana12 have all provided some clues about its function. It has become apparent from these mutants that there is no essential role for SQDG in photosynthetic bacteria or in eukaryotic cells containing chloroplasts because all mutants are capable of photoautotrophic growth. The effects on photosynthesis are subtle under normal growth conditions. However, if these SQDG-deficient null mutants are starved for P, they cease growth much earlier than the corresponding wild-type12,42,43. Thus, SQDG has been considered to be of conditional importance in these organisms, including plants. In addition, SQDG and PG contents are known to be maintained by reciprocally balancing their levels with changes in P availability, which is considered to contribute to the maintenance of the negative charge in thylakoid membranes12,45. However, among the three SQDG-deficient mutants of A. thaliana, the severe growth defect under the P-limited condition was observed only in the sqd2 mutant (Fig. 4b and Supplementary Fig. S11)12, and no studies have investigated the differences in the lipid profiles of these SQDG-deficient mutants11,12,17.
Our present results unexpectedly revealed that the P-starved sqd2 mutant of A. thaliana lacks not only SQDG but also a novel anionic glycolipid GlcADG, which results in a severe phenotype in this mutant that is not observed in the other two SQDG-deficient mutants analysed in this study. In addition, under P limitation, the ugp3 and sqd1 mutants accumulated slightly higher levels of GlcADG than the wild-type and showed no apparent growth defects. These results collectively indicate the following: (1) GlcADG is a novel molecule importantly involved in the protection against P-depletion stress in plants; (2) the growth defect of P-starved sqd2 is attributable to the deficiency of the two anionic glycolipids SQDG and GlcADG; and (3) moderate increases in GlcADG can compensate for the SQDG deficiency, as shown in the sqd1 and ugp3 mutants. Our results reinforce the importance of plant SQDG synthase and anionic glycolipids synthesized by SQDG synthase during growth under P-limited conditions.
The details of the biosynthesis of GlcADG remain unknown except for the fact that SQDG synthase is required for GlcADG biosynthesis. We propose a possible biosynthetic scheme for this glycolipid in Fig. 4. However, our lipidomic data using a few SQDG-deficient mutants do not deny the possibility of interconversion of SQDG and GlcADG. Other types of experiments, such as flux analysis, will be important in understanding the metabolic flow underlying the accumulation of these two anionic glycolipids. It is also postulated that the inducible synthesis of GlcADG under P starvation entails increased synthesis of an unidentified sugar donor, presumably UDP-GlcA, in chloroplasts. Our current study suggests the possibility of the presence of an uncharacterized biosynthetic pathway or transportation mechanism for UDP-GlcA in chloroplasts. The synthesis of GlcADG by dehydrogenation of monoglucosyldiacylglycerol, found in several plants46, is also possible.
In P-starved sqd2 mutants, the levels of PI and PE were significantly elevated, which indicates that the complete deficiency of anionic glycolipids causes the de-regulation of not only PG but also other phospholipids, such as PI and PE. Thus, levels of these phospholipids (PI and PE) and GlcADG are also reciprocally maintained in plant cells under P-limited conditions (Fig. 6), as a similar relationship can be found between SQDG and PG, suggesting some shared functions among GlcADG, PI and PE under P-limited conditions. Levels of 34:3 PI and 34:3 PE were also slightly elevated in sqd1 and ugp3 mutants compared with wild-type even under P sufficiency (Supplementary Fig. S8). Thus, these phospholipids may also generally contribute to the maintenance of proper membrane properties in plants even under P sufficiency, and their roles may become more important when plants are deficient in two anionic glycolipids under P-depleted conditions.
An abbreviated biosynthetic scheme for the major plant polar glycerolipids is represented along with the results of the current report. Previous research on several SQDG-deficient mutants suggested that the levels of SQDG and PG are reciprocally regulated (orange line), while our current report suggests that a similar reciprocal regulation also can be found among GlcADG, PI and PE (red lines). The levels of SQDG, PI and PE might also be reciprocally maintained.
Currently, contributions to the compensation of the deficiency of anionic glycolipids are likely to be significant in the order of PG>PI>PE, although it remains unknown whether this order is because of the difference in chemical properties or the difference in the organelle where they are mainly synthesized, that is, chloroplastic PG is synthesized in chloroplasts47, such as SQDG and presumably GlcADG, whereas PI and PE are mainly synthesized in the endoplasmic reticulum (Fig. 6). We have never determined where GlcADG is synthesized and accumulates, but GlcADG is likely synthesized in chloroplasts because SQDG synthase is localized in the chloroplast inner membrane37.
The idea that GlcADG accumulates in chloroplasts is plausible considering the localization of SQDG synthase, although we cannot exclude the possibility that somehow GlcADG is exported from chloroplasts to extraplastidic membranes such as DGDG6,8. Given that GlcADG accumulates in extraplastidic space, we might be able to determine the reason why the levels of GlcADG, PI and PE are reciprocally regulated in P-starved plants. In addition, we cannot deny the possibility that levels of PI and PE in the chloroplast membrane are elevated in sqd2 mutants under P deficiency even though PE is usually not abundant in chloroplasts48. Further research to determine the localization of GlcADG accumulation will help us to understand this phenomenon. A study on the bacterium B. diminuta also investigated the replacement of acidic phospholipids by GlcADG upon P limitation49, suggesting a functional convergence of lipid remodelling involving the accumulation of GlcADG across kingdoms. No information regarding the genes involved in GlcADG biosynthesis in B. diminuta is currently available.
Lipid remodelling in membrane systems under P-limited conditions is considered to contribute to sustaining the level of cellular inorganic Pi by modifying the P allocation in the cell. As described above, lipid remodelling is severely de-regulated in a P-starved sqd2 mutant, but there is no significant decrease in free Pi in this mutant (Supplementary Fig. S13). These results suggest that GlcADG is not essential to maintain the free Pi level in P-starved plants but that it is likely to contribute to the control of the membrane environment to ensure the proper functioning of organelles, including chloroplasts, in P-limited, unfavourable environments.
In addition, this study suggests that plants have developed an alternative mechanism to maintain a negative membrane charge by synthesizing uronic acid-containing lipids free of sulphur, which is also an essential macronutrient requiring significant energy for its assimilation50. Genome sequence information on organisms capable of synthesizing SQDG has revealed that the two genes coding UDP-SQ synthase and SQDG synthase evolved at different times, and while many genomes show readily identifiable UDP-SQ synthase orthologs, SQDG synthases were differently recruited in cyanobacteria/plants versus non-cyanobacteria and archaea51. The involvement of plant SQDG synthase in different anionic glycolipids might be related to its ancestral function, which might still be retained in plants to allow them adapt to unfavourable surroundings.
Methods
Chemicals
Primuline, (trimethylsilyl)diazomethane (2.0 M in diethyl ether), and 1,2-didecanoyl-sn-glycero-3-phosphocholine were obtained from Sigma-Aldrich, and polypropylene glycol 2000 (PPG-2000) was obtained from Alfa Aesar. Glucuronic acid was purchased from Tokyo Chemical Industry. Unless stated otherwise, all other chemicals were purchased from Wako Pure Chemical Industries.
Nuclear magnetic resonance
1H NMR spectra were recorded in CDCl3 supplemented with a drop of D2O for H-D exchange. Other NMR spectra (13C, COSY, HMQC, HMBC and DEPT) were recorded in absolute CDCl3. Tetramethylsilane was used for the internal standard.
Plant materials
A. thaliana—Seeds of T-DNA insertion lines for sqd2-1 (SALK_070595) and sqd2-2 (SALK_139798) were purchased from ABRC at Ohio State University52. The T-DNA insertion sites of these lines were confirmed by sequencing PCR fragments (Supplementary Fig. S16). A PCR fragment at the left border of the T-DNA of sqd2-1 was amplified using LBa1 and SALK_070595_Fw, and the right border of the T-DNA of sqd2-2 was amplified using SALK_070595_Fw and RBa1 (Supplementary Table S1). The sqd1, ugp3-1 and ugp3-2 mutants were isolated in our previous study based on the results of genome PCR17 and used again in this research. Seeds of the sqd2 mutant in the Ws-0 background (provisionally named sqd2-3 in this study) and its wild-type were kind gifts from Professor Benning12. Surface-sterilized seeds were sown on agar-solidified Murashige and Skoog medium53 containing 1% (w/v) sucrose at 22 °C under 16 h light/8 h dark cycles. Photon flux density was 50 μmol m–2 sec–1. The seedlings were kept on agar for 14 days before transfer to plates with controlled phosphate (Pi) concentrations. For P-limitation experiments, sterile Arabidopsis medium54 was used but at half concentration and containing 0.8% (w/v) agar, 20 mM MES (pH 6.0) and different concentrations of KH2PO4 as indicated7. For lipid analysis, leaves were harvested 6 h after the onset of the light phase, frozen in liquid nitrogen and stored at −80 °C until lipid extraction.
Rice—Dehulled seeds of O. sativa L. cv. Nipponbare were sterilized and germinated on agar-solidified Murashige and Skoog medium at 28 °C under a 16 h light/8 h dark cycle. To expose plants to P-limiting stress, 5-day-old seedlings were transplanted and hydroponically cultured in the P-controlled medium used for the stress treatment of A. thaliana. The culture medium was changed every week.
Lipidomics by HILIC-MS
Plant lipids were extracted following the method of Bligh and Dyer55 with slight modifications26, and directly subjecting the extract to LC-MS analysis. The lipidomic data were recorded on an LC-MS-IT-TOF mass spectrometer using electrospray ionization, combined with an LC-20AD HPLC system (Shimadzu)26. Parameters for MS were as follows: scan range, m/z 150–1,600; loop time, 0.6 s; CDL and heat block temperature, 160 °C; nebulizer gas (N2), 1.5 l min–1. More details are found in our previous report26 and metadata in ‘Supplementary Data Set.xls’.
Peak picking and alignment were performed using the Profiling Solution software (version 1.0.76.0; Shimadzu). The profiling solution parameters were as follows: ion m/z tolerance, 20 mDa; ion RT tolerance, 0.1 min; ion intensity threshold, 2e4; detect isomer valley, 20%; allow some ion without isotope peak, ON26. The data matrix containing ~480 ions consisting of negatively detected ions was exported from the profiling solution software and normalized relative to the intensity of the [M+HCO2]− ion of the internal standard (1,2-didecanoyl-glycero-3-phosphocholine). Annotation was based on the theoretical m/z values of each lipid species and the retention time of identified compounds with the same polar head26.
Statistics for the analysis of lipidomic data
Normalized LC-MS data were pareto-scaled and subjected to OPLS-DA using SIMCA-P (version 11.0.0.0; Umetrics). Tukey’s test and Welch’s t-test were performed using their respective functions (TukeyHSD and t-test) in the statistical software R version 2.14.0 ( http://www.r-project.org/). HCL and jackknife testing were performed on MultiExperiment Viewer (MeV) version 4.8 ( http://www.tm4.org/mev/). For HCL, Pearson’s correlations were used to analyse the distance metric. Average linkage clustering was selected for the linkage method. The data set used for HCL is available as ‘Supplementary Data Set.xls’. Metadata also can be found in the same file.
Preparation of GlcADG from A. thaliana and B. diminuta
A. thaliana plants (Col-0 accession, 340 g fresh weight) grown under P deficiency as described above were extracted using the same protocol to prepare samples for lipidomic analysis. The resulting lipid extracts (4.1 g) was loaded onto a silica gel column (silica gel 60 N, spherical, neutral; 25-cm long, 7-cm i.d.; Kanto Chemical) equilibrated with chloroform. The column was washed with chloroform (3 l), chloroform–acetone (1:1, v/v, 3.6 l), acetone (1 l), acetone–methanol (1:1, v/v, 1 l) and methanol (1 l). The acetone–methanol fractions containing UK1 were subjected to preparative thin-layer chromatography on Silica Gel 60 plates containing no fluorescent indicator (Merck) using chloroform–methanol–H2O (65:25:4, v/v/v). After spraying a primuline solution, lipids were detected under UV light. The fluorescent spots containing UK1 were scraped off and further subjected to preparative thin-layer chromatography with a chloroform–methanol–acetic acid solution (75:25:3, v/v/v) to obtain UK1 (1, 9.6 mg). MS/MS of all detectable UK1 molecules, are found in Fig. 2 and Supplementary Figs S2,S3.
B. diminuta ATCC 11568 (Strain JCM 2788) was provided by RIKEN BRC (Wako, Japan). The strain was rehydrated according to the provider’s instructions and cultivated with continuous rotation at 180 r.p.m. in lysogeny broth containing PPG-2000 (50 p.p.m.) as antifoam. After a 48-h incubation at 30 °C, the cells were collected by centrifugation. The dried cells (4.43 g) were extracted with mixtures of chloroform, methanol and diethyl ether to yield crude lipid extracts (320 mg) as previously reported56,57. The extracts were separated to purify GlcADG (17.8 mg) using the same protocol described above for the purification of UK1. MS and MS/MS of the bacterial GlcADG are found in Supplementary Fig. S4.
Preparation of the methyl ester of GlcADG
GlcADG from B. diminuta and UK1 from A. thaliana were methylated with (trimethylsilyl)diazomethane as previously described58. The 1H and 13C NMR spectra and key HMBCs of bacterial GlcADG are shown in Supplementary Figs S17–S19.
Methyl ester of GlcADG from B. diminuta.1H NMR (500 MHz, CDCl3) δ 0.84–0.93 (6 H, m, –CH2CH3), 1.19–1.45(ca 36 H, m, –CH2–), 1.54–1.66 (4 H, m, –OCOCH2CH2–), 1.93––2.10 (5 H, m, –CH=CHCH2–), 2.25–2.39 (4 H, m, –OCOCH2R), 3.57 (1 H, dd, J=3.7, 9.5 Hz, H-2′), 3.67 (1 H, dd, J=5.7, 10.9 Hz, H-3a), 3.72 (1 H, triplet-like dd, J=8.9, 9.5 Hz, H-4′), 3.78 (1 H, triplet-like dd, J=8.9, 9.5 Hz, H-3′), 3.83 (3 H, s, CH3O-6′), 3.90 (1 H, dd, J=4.6, 10.9 Hz, H-3b), 4.15 (1 H, dd, J=5.7, 11.9 Hz, H-1a), 4.16 (1 H, d, J=9.5 Hz, H-5′), 4.41 (1 H, dd, J=4.2, 11.9 Hz, H-1b), 4.94 (1 H, d, J=3.7 Hz, H-1′), 5.26 (1 H, m, H-2), 5.33–5.36 (2 H, m, –CH=CH–); 13C NMR (125 MHz, CDCl3) δ 14.1 (–CH2CH3), 22.66–22.70 (–CH2CH2CH3), 24.9 (–OCOCH2CH2CH2–), 27.2 (–CH=CHCH2CH2–), 28.7–31.9 (–CH2CH2CH2–), 34.1–34.2 (–OCOCH2CH2–), 52.9 (CH3O-6′), 61.9 (C-1), 67.1 (C-3), 69.6 (C-2), 70.6 (C-5′), 71.5 (C-2′, C-4′), 73.6 (C-3′), 99.1 (C-1′), 129.8 (–CH=CH–), 130.0 (–CH=CH–), 170.5 (C6′), 173.2 (–OCOR of fatty acid), 173.5 (–OCOR of fatty acid).
Methyl ester of UK1 from A. thaliana.1H NMR (500 MHz, CDCl3) δ 0.85–0.93 (ca 4 H, m, –CH2CH3), 0.95–1.02 (ca 3 H, –CH=CHCH2CH3), 1.2–1.4 (ca 40 H, m, –CH2–), 1.5–1.7 (ca 13 H, m, –OCOCH2CH2–), 2.01–2.13 (ca 6 H, m, –CH=CHCH2–), 2.27–2.37 (ca 4 H, m, –OCOCH2R), 2.74–2.84 (ca 4 H, m, –CH=CHCH2CH=CH–), 3.58 (1 H, dd, J=3.7, 9.5 Hz, H-2′), 3.68 (1 H, dd, J=5.7, 10.9 Hz, H-3a), 3.72 (1 H, triplet-like dd, J=8.9, 9.5 Hz, H-4′), 3.79 (1 H, triplet-like dd, J=8.9, 9.5 Hz, H-3′), 3.84 (3 H, s, CH3O-6′), 3.91 (1 H, dd, J=4.6, 10.9 Hz, H-3b), 4.14 (1 H, dd, J=5.7, 12.0 Hz, H-1a), 4.17 (1 H, d, J=9.5 Hz, H-5′), 4.42 (1 H, dd, J=4.3, 12.0 Hz, H-1b), 4.95 (1 H, d, J=3.7 Hz, H-1′), 5.23–5.29 (ca 1 H, m, H-2), 5.29–5.44 (ca 8 H, m, -CH=CH-); 13C NMR (125 MHz, CDCl3) δ 14.06–14.3 (-CH2CH3), 20.5 (-CH=CHCH2CH3), 22.6–22.7 (-CH2CH2CH3), 24.8 (-OCOCH2CH2CH2-), 25.5–25.6 (-CH=CHCH2CH=CH-), 27.2 (-CH=CHCH2CH2-), 29.1–31.9 (-CH2CH2CH2-), 34.1–34.2 (-OCOCH2CH2-), 52.8 (CH3O-6′), 61.9 (C-1), 67.1 (C-3), 69.7 (C-2), 70.6 (C-5′), 71.5 (C-2′, C-4′), 73.6 (C-3′), 99.1 (C-1′), 127.1–132.0 (olefinic carbons of fatty acids), 170.4 (C6′), 173.1 (-OCOR of fatty acid), 173.5 (-OCOR of fatty acid).
Quantification of inorganic Pi in plants
Leaves (30 mg) were frozen in liquid nitrogen and homogenized using a Mixer Mill MM300 (Retsch, Haan, Germany) for 2 min at 15 Hz with a zirconium bead and then extracted with 50% (v/v) aqueous methanol (300 μl) followed by centrifugation at 12,000 g. The milling process was carried out under cryogenic conditions (the grinding jars were cooled with liquid nitrogen before each milling process to ensure a low milling temperature and keep the plants frozen). The measurement of Pi content in the supernatants was accomplished by forming a phosphomolybdate complex and in turn reducing it to a molybdenum blue complex59.
Additional information
How to cite this article: Okazaki, Y. et al. A new class of plant lipid is essential for protection against phosphorus depletion. Nat. Commun. 4:1510 doi: 10.1038/ncomms2512 (2013).
References
Raghothama K. G. Phosphate transport and signaling. Curr. Opin. Plant Biol. 3, 182–187 (2000).
Vance C. P. Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol. 127, 390–397 (2001).
Cordell D., Rosemarin A., Schroder J. J., Smit A. L. Towards global phosphorus security: A systems framework for phosphorus recovery and reuse options. Chemosphere 84, 747–758 (2011).
Gilbert N. Environment: the disappearing nutrient. Nature 461, 716–718 (2009).
Van Kauwenbergh S. J. World phosphate rock reserves and resources International Fertilizer Development Center (2010).
Andersson M. X., Stridh M. H., Larsson K. E., Liljenberg C., Sandelius A. S. Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 537, 128–132 (2003).
Härtel H., Dörmann P., Benning C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl Acad. Sci. USA 97, 10649–10654 (2000).
Jouhet J. et al. Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J. Cell Biol. 167, 863–874 (2004).
Benning C., Ohta H. Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 280, 2397–2400 (2005).
Andersson M. X., Larsson K. E., Tjellström H., Liljenberg C., Sandelius A. S. Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J. Biol. Chem. 280, 27578–27586 (2005).
Essigmann B., Guler S., Narang R. A., Linke D., Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 95, 1950–1955 (1998).
Yu B., Xu C., Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl Acad. Sci. USA 99, 5732–5737 (2002).
Kobayashi K. et al. Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 57, 322–331 (2009).
Kobayashi K., Masuda T., Takamiya K., Ohta H. Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J. 47, 238–248 (2006).
Gaude N., Nakamura Y., Scheible W. R., Ohta H., Dormann P. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 56, 28–39 (2008).
Awai K. et al. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 98, 10960–10965 (2001).
Okazaki Y. et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21, 892–909 (2009).
Kelly A. A., Dormann P. DGD2, an arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 277, 1166–1173 (2002).
Cruz-Ramirez A., Oropeza-Aburto A., Razo-Hernandez F., Ramirez-Chavez E., Herrera-Estrella L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl Acad. Sci. USA 103, 6765–6770 (2006).
Nakamura Y. et al. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280, 7469–7476 (2005).
Nakamura Y. et al. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl Acad. Sci. USA 106, 20978–20983 (2009).
Cheng Y. et al. Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 66, 781–795 (2011).
Bari R., Datt Pant B., Stitt M., Scheible W. R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999 (2006).
Narise T. et al. Involvement of auxin signaling mediated by IAA14 and ARF7/19 in membrane lipid remodeling during phosphate starvation. Plant Mol. Biol. 72, 533–544 (2010).
Li M., Welti R., Wang X. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142, 750–761 (2006).
Okazaki Y., Kamide Y., Hirai M. Y., Saito K. Plant lipidomics based on hydrophilic interaction chromatography coupled to ion trap time-of-flight mass spectrometry. Metabolomics doi:10.1007/s11306-011-0318-z (2011).
Wiklund S. et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 80, 115–122 (2008).
Hölzl G., Dörmann P. Structure and function of glycoglycerolipids in plants and bacteria. Prog. Lipid Res. 46, 225–243 (2007).
Eichenberger W., Gribi C. Diacylglycerol-α-D-glucuronide from Ochromonas danica (Chrysophyceae). J. Plant Physiol. 144, 272–276 (1994).
Eichenberger W., Gribi C. Lipids of Pavlova lutheri: cellular site and metabolic role of DGCC. Phytochemistry 45, 1561–1567 (1997).
Wilkinson S. G. Lipids of Pseudomonas diminuta. Biochim. Biophys. Acta 187, 492–500 (1969).
Batrakov S. G., Nikitin D. I., Sheichenko V. I., Ruzhitsky A. O. Unusual lipid composition of the gram-negative, freshwater, stalked bacterium Caulobacter bacteroides NP-105. Biochim. Biophys. Acta 1347, 127–139 (1997).
Fontaine T. et al. Characterization of glucuronic acid containing glycolipid in Aspergillus fumigatus mycelium. Carbohydr. Res. 344, 1960–1967 (2009).
Browse J., Warwick N., Somerville C. R., Slack C. R. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the '16:3' plant Arabidopsis thaliana. Biochem. J. 235, 25–31 (1986).
Heinz E., Roughan P. G. Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol. 72, 273–279 (1983).
Sanda S., Leustek T., Theisen M. J., Garavito R. M., Benning C. Recombinant Arabidopsis SQD1 converts UDP-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 276, 3941–3946 (2001).
Tietje C., Heinz E. Uridine-diphospho-sulfoquinovose: diacylglycerol sulfoquinovosyltransferase activity is concentrated in the inner membrane of chloroplast envelopes. Planta 206, 72–78 (1998).
Seifert U., Heinz E. Enzymatic characteristics of Udp-sulfoquinovose - diacylglycerol sulfoquinovosyltransferase from chloroplast envelopes. Bot. Acta 105, 197–205 (1992).
Heinz E. et al. Synthesis of different nucleoside 5′-diphospho-sulfoquinovoses and their use for studies on sulfolipid biosynthesis in chloroplasts. Eur. J. Biochem. 184, 445–453 (1989).
Shimojima M. Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 50, 234–239 (2011).
Benning C. Biosynthesis and Function of the Sulfolipid Sulfoquinovosyl Diacylglycerol. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 53–75 (1998).
Benning C., Beatty J. T., Prince R. C., Somerville C. R. The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc. Natl Acad. Sci. USA 90, 1561–1565 (1993).
Güler S., Seeliger A., Härtel H., Renger G., Benning C. A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J. Biol. Chem. 271, 7501–7507 (1996).
Sato N., Sonoike K., Tsuzuki M., Kawaguchi A. Impaired photosystem II in a mutant of Chlamydomonas reinhardtii defective in sulfoquinovosyl diacylglycerol. Eur. J. Biochem. 234, 16–23 (1995).
Yu B., Benning C. Anionic lipids are required for chloroplast structure and function in Arabidopsis. Plant J. 36, 762–770 (2003).
Jamieson G. R., Reid E. H. Sugar components of galactosyl diglycerides from green plants. Phytochemistry 15, 135–136 (1976).
Xu C. et al. The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol. 129, 594–604 (2002).
Block M. A., Douce R., Joyard J., Rolland N. Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth. Res. 92, 225–244 (2007).
Minnikin D. E., Abdolrahimzadeh H., Baddiley J. Replacement of acidic phosphates by acidic glycolipids in Pseudomonas diminuta. Nature 249, 268–269 (1974).
Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184 (2011).
Benning C., Gravito R. M., Shimojima M. in Sulfur Metabolism in Phototrophic Organisms (Advances in Photosynthesis and Respiration) (eds Hell R., Dahl C., Knaff D. B., Leustek T. ) Ch. 10, 185–200Springer (2008).
Alonso J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003).
Murashige T., Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497 (1962).
Estelle M. A., Somerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206 (1987).
Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Wilkinson S. G. Glycosyl diglycerides from Pseudomonas rubescens. Biochim. Biophys. Acta 164, 148–156 (1968).
Wilkinson S. G. Studies on the cell walls of Pseudomonas species resistant to ethylenediaminetetra-acetic acid. J. Gen. Microbiol. 54, 195–213 (1968).
Hashimoto N., Aoyama T., Shioiri T. New methods and reagents in organic synthesis.14. A simple efficient preparation of methyl esters with trimethylsilyldiazomethane (TMSCHN2) and its application to gas chromatographic analysis of fatty acids. Chem. Pharm. Bull 29, 1475–1478 (1981).
Ames B. N. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8, 115–118 (1966).
Murphy R. C. in Handbook of Lipid Research (ed Snyder F. ) vol. 7 Chapter 7, 223–226Plenum Press (1993).
Acknowledgements
We are grateful to Professor Christoph Benning (Michigan State University) for kindly donating the sqd2 mutant seeds (provisionally named as sqd2-3) and the wild-type A. thaliana seeds (Ws-0 accession). We also thank our colleague Dr Ryo Nakabayashi (RIKEN Plant Science Centre) for his advice regarding the NMR experiments. The stock of B. diminuta was provided by the Japan Collection of Microorganisms, RIKEN BRC (Wako, Japan), which is participating in the National BioResource Project, MEXT, Japan. This work was supported in part by grant aids from the Strategic International Research Cooperative Programme of Japan Science Technology (Metabolomics for a Low Carbon Society, JST-NSF) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This research was also supported by the Japan Advanced Plant Science Network.
Author information
Authors and Affiliations
Contributions
Y.O. performed the metabolite profiling of plants. H.O., T.N., Y.K. and M.Y.H. performed the molecular biology experiments. Makoto K. and Miyako K. performed the GC–MS analysis. S.S. and T.A. performed the NMR experiments. Y.O. and K.S. designed the experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S19, Supplementary Table S1 and Supplementary References (PDF 1243 kb)
Supplementary Dataset 1
Supplementary Dataset 1 (XLS 151 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Okazaki, Y., Otsuki, H., Narisawa, T. et al. A new class of plant lipid is essential for protection against phosphorus depletion. Nat Commun 4, 1510 (2013). https://doi.org/10.1038/ncomms2512
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms2512
This article is cited by
-
NUTCRACKER-overexpressing Arabidopsis thaliana grows better under phosphate starvation
Plant Growth Regulation (2023)
-
The regulation of sulfolipids under sulfur starvation
Plant Molecular Biology (2023)
-
Metabolomics analysis of milk thistle lipids to identify drought-tolerant genes
Scientific Reports (2022)
-
Membrane lipid phosphorus reusing and antioxidant protecting played key roles in wild soybean resistance to phosphorus deficiency compared with cultivated soybean
Plant and Soil (2022)
-
Adaptive strategies of plants to conserve internal phosphorus under P deficient condition to improve P utilization efficiency
Physiology and Molecular Biology of Plants (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.