Abstract
Molecular self-assembly with DNA offers a route for building user-defined nanoscale objects, but an apparent requirement for magnesium in solution has limited the range of conditions for which practical utility of such objects may be achieved. Here we report conditions for assembling templated multi-layer DNA objects in the presence of monovalent ions, showing that neither divalent cations in general or magnesium in particular are essential ingredients for the successful assembly of such objects. The percentage of DNA strands in an object that do not form thermally stable double-helical DNA domains (Tm>45 °C) with the template molecule correlated with the sodium requirements for obtaining folded objects. Minimizing the fraction of such weakly binding strands by rational design choices enhanced the yield of folding. The results support the view that DNA-based nanodevices may be designed and produced for a variety of target environments.
Similar content being viewed by others
Introduction
Molecular self-assembly with DNA1,2 enables building custom-shaped nanometer-precise objects with addressable chemical features3,4,5,6,7,8. DNA origami9 is a successful design strategy that entails the folding of a long-template DNA strand ('scaffold') into target shapes that are encoded in the sequences of a set of short DNA strands ('staples'). DNA origami has enabled the construction of sophisticated single-layered DNA patterns and containers10,11,12,13,14,15 along with multi-layer objects16,17,18,19,20 that consist of multiple layers of DNA double helices packed in parallel. DNA origami shapes have been tailored for custom applications, among them are nanotubes for NMR-based structural biology21,22, DNA-hybrid-devices for electronics23,24, logic-gated capsules for cell-specific molecular delivery25, optically active polarizers that may create metafluids26 and gatekeepers for single-molecule sensing with nanopores27.
The great majority of these objects have been synthesized using a method of attractive simplicity, namely by thermal annealing of one-pot mixtures containing all required DNA strands (Fig. 1a). However, such reactions often result in a complex mixture containing fully folded objects and also by-products such as aggregates and unfolded or partially folded objects16. The success of assembly hinges on many aspects to be considered during object design and practical assembly, including structural details, choice of sequences, choice of annealing protocol, choice of concentrations and choice of co-factors. The divalent cation magnesium has long been a co-factor in DNA-based molecular self-assembly reactions (with few exceptions28), owing to its efficacy in screening inter-helical repulsion and stabilizing the stacked form of DNA Holliday junctions29. Assembling multi-layer DNA origami objects was found to require precisely calibrated magnesium chloride concentrations; additional monovalent cations antagonized folding16. However, magnesium is often not desirable: magnesium promotes DNA degradation, it can trigger unwanted enzymatic activity30, it may affect the fluorescence brightness of dye molecules31 and precipitate inorganic nanoparticles32,33.
(a) Schematic illustration of templated molecular self-assembly with DNA origami using one-pot thermal annealing. (b,d) Photographs of ethidium-bromide-stained 2% agarose gels with products obtained from 12-day-long thermal annealing (Supplementary Tables S5) of a multi-layer DNA object in magnesium chloride (MgCl2) (b) and in sodium chloride (NaCl) (d). Saturated high-mobility bands are caused by excess staple strands. The object is designed as 42 DNA double helices packed in parallel on a honeycomb-type lattice. Arrows marked with 'u' indicates unfolded species, whereas 'f' indicates the folded particle species. M, 1 kb marker; ref, scaffold DNA. (c,e) Representative average negative-stain TEM micrographs of folded particles. Particle micrographs were obtained by imaging unpurified folding products directly after thermal annealing in (c) 20 mM magnesium chloride and (e) 2.4 M sodium chloride. Flat view: sum of 300 particles each, side view: sum of 50 particles each. See Supplementary Fig. S2 for individual particles. Scale bar, 20 nm.
Here, conditions are reported for assembling multi-layer DNA objects in the absence of magnesium but in the presence of monovalent sodium, showing that neither divalent cations in general or magnesium in particular are essential requirements for assembling such objects and conferring practical advantages for applications that are not compatible with magnesium or divalent cations.
Results
Self-assembly in the presence of either magnesium or sodium
For one set of experiments, we designed a multi-layer DNA origami nanostructure in which 42 DNA double helices were bundled in honeycomb-type lattice packing to form a rectangular brick-shaped object. As a positive control, we analysed the folding behaviour of this object in magnesium-chloride-containing reactions, at concentrations ranging from 0 to 32 mM MgCl2 in steps of 2 mM. Note that the concentration of free magnesium ions was reduced due to the presence of 1 mM EDTA in solution. The reactions were subjected to slow thermal annealing from 65 to 25 °C over periods of up to 23 days. The reaction products were analysed using agarose gel electrophoresis (Fig. 1b). The reaction that did not contain magnesium produced a smear in the gel that points to single-stranded scaffold DNA degradation. For the reactions with magnesium chloride concentrations up to 6 mM, we observed a single band that corresponded to unstructured particles or partially structured particles, in accordance with previous work16. A faster migrating species emerged for higher magnesium concentrations that corresponded to correctly folded particles, which we verified using negative-stain transmission electron microscopy (TEM). When using shorter annealing ramps, we found that the concentration range for which unstructured and folded species coexist broadened toward higher magnesium concentrations (Supplementary Fig. S1). Increasingly high assembly barriers when decreasing the magnesium chloride concentration may explain the increasing coexistence regime for shorter annealing ramps. A concentration of 8 mM magnesium chloride appeared minimally required for full assembly of the 42-helix bundle.
We repeated the thermal annealing experiments with the 42-helix bundle object with sodium chloride instead of magnesium chloride in the reaction mixtures. We screened 100-fold larger concentrations than for magnesium chloride, from 0 to 3.4 M sodium chloride in steps of 0.2 M. The reaction products were again analysed using agarose gel electrophoresis (Fig. 1d). For concentrations up to 1 M sodium chloride, a slow band was observed that reflected the presence of unstructured particles, whereas at 1.2 M sodium chloride, the emergence of a faster migrating species reflected the additional presence of folded particles. The folded species became enriched towards higher sodium chloride concentrations. In contrast to assembly in the presence of magnesium, coexistence of folded and unstructured species prevailed over a wide concentration range from 1.2 M up to 3.4 M sodium chloride even in 23-day long annealing reactions. The overall lane intensities remained comparable up to 3.4 M NaCl.
To compare the shape of folded objects formed in the presence of either magnesium or sodium, we employed negative-stain TEM and image processing (Fig. 1c,e; Supplementary Fig. S2). In micrographs prepared by classifying and averaging distinct individual particle views, we found that objects assembled in magnesium chloride versus sodium chloride were highly similar and agreed both favourably with the expected shape (Fig. 1c,e). A gel electrophoretic analysis of products obtained from reactions run with either magnesium or sodium, but adjusted to identical ionic strength post folding (Supplementary Fig. S3), showed identical gel mobility of the folded species, thus supporting the interpretation that the folded objects were similar in shape.
Shape- and design-specific sodium requirements
We designed a panel of ten additional multi-layer DNA origami objects (Fig. 2a) including 6, 8, 10, 12, 18 and 24 helix bundles in honeycomb-lattice packing, a 100-helix bundle in square-lattice packing and three additional versions of the 42-helix bundle. We screened the assembly behaviour of all of these structures upon thermal annealing in the presence of varying concentrations of either magnesium chloride (as a positive control for folding) or sodium chloride and analysed the reaction products using gel electrophoresis as described above for Fig. 1. All structures assembled successfully in both types of electrolytes except for the 100-helix bundle, which only formed in the presence of magnesium.
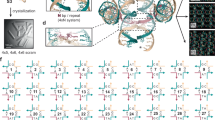
(a) Cylinder models of 6, 8, 10, 12, 18, 24, 42 and 100 helix (hlx) bundle objects. Four different versions of the 42-helix bundle object were designed. See Supplementary Fig. S5 for design details and Supplementary Data 1 for DNA sequences for all objects. (b,c) Grey triangles: minimally required magnesium chloride concentrations for obtaining fully folded particles of each of the indicated DNA objects in slow one-pot thermal annealing. Red circles: minimally required sodium chloride concentrations for obtaining fully folded particles in slow one-pot thermal annealing. See Supplementary Fig. S1 for gel electrophoretic data. Blue squares: fraction of DNA staple strands in each object that do not form double-helical DNA domains with the scaffold strand with melting temperatures above 45 °C (methods).
We determined the minimally required concentration of either magnesium or sodium ions for emergence of fully folded particles for each object tested (Fig. 2b,c, see also Supplementary Fig. S1 for gel electrophoretic data). We found that thin objects, such as the six-helix bundle, formed in the presence of 200 mM sodium, whereas thicker objects such as the 24-helix bundle fully formed only in the presence of molar concentrations of sodium (1 M). The sodium requirements grew with increasing thickness of the objects, but also varied for design variants of the same global shape as observed for the four versions of the 42-helix bundle object.
The four versions of the 42-helix bundle object served as test cases for the influence of decisions taken during design (here: staple backbone breaking rules) on folding success in the presence of sodium. All four versions of the 42-helix bundle used staple strands with an average length of 42 nucleotides derived from identical scaffold sequence permutations. In rule 1 (Fig. 3a), backbone nicks were placed predominantly in dsDNA segments between crossovers longer than 7 bp and positioned 3 or 4 bp away from crossovers. In 7-bp segments, nicks were positioned 3 and 4 bp away from crossovers. Rule 2 (Fig. 3a) was similar to rule 1 except that nicks were placed only 2 bp away from the nearest crossover. In rule 3 (Fig. 3a), backbone nicks were placed directly at crossover positions such that each staple strand wherever possible, forms one, but only one, 14-bp dsDNA domain with the scaffold. The rule 3 version, therefore, contained also half crossovers. Rule 4 (Fig. 3a) was a partial negation of rule 1; backbone nicks were never placed in dsDNA segments longer than 7 bp. In 7-bp segments, nicks were positioned 3 and 4 bp away from crossovers. The four different design rules resulted in distinct distributions of dsDNA domain lengths (Fig. 3b) that are formed between staple strands and scaffold within the 42-helix bundle object.
(a) Schematic illustration of four backbone nick distribution rules. Thick solid lines represent scaffold strand. The horizontal crossover spacing is 7 bp for honeycomb-type packing. (b) The distribution of length of dsDNA domains formed by staple strands with the scaffold strand resulting when applying backbone nick rules 1 to 4 to a 42-helix bundle object in honeycomb-type packing. (c) Left panel (middle panel): photograph of a ethidium-bromide-stained 2% agarose gel on which folding products of 42-helix bundle versions designed according to rules 1 to 4 obtained after 1.5-day-long (12-day-long) thermal annealing in the presence of 2.4 M NaCl were electrophoresed. M, 1 kb marker; ref, scaffold DNA. (d) As in (c) left panel, but for reactions run in the presence of 20 mM MgCl2. M, 1 kb marker; ref, scaffold DNA.
The four versions of the 42-helix bundle exhibited a markedly different assembly behaviour in the presence of sodium chloride. For example, the object designed according to rule 4 was the only version that fully formed in short 1.5-day-long annealing (Fig. 3c), while all design versions gave folded products after 12-day-long annealing (Fig. 3c) in the presence of 2.4 M sodium. Note that the object designed according to rule 4 had the highest mobility, suggesting strongest compaction. For 12-day annealing, the yield of folding was also best for the object designed according to rule 4 with negligible by-products and approaches 100%, as measured by product band intensity versus total lane intensity excluding the excess staple band (Fig. 3c). The objects designed according to rules 1 to 4 could be ordered according to minimally required sodium concentrations (Fig. 2c). The rule 1 object required the most sodium (1.6 M), whereas the rule 4 object required the least sodium (1 M).
In the presence of 20 mM magnesium instead of sodium, all versions of the 42-helix bundle fully formed in short 1.5-day-long annealing reactions (Fig. 3d). In such reactions, all versions yielded fully folded particles, but rule 3 and rule 4 reactions gave the best results with fewest by-products. In another recent study34, various backbone nick distribution rules were tested for their ability to increase the yield of assembly in the presence of magnesium chloride reaction. The presence of a 14-bp-long 'seed' dsDNA domain formed by each staple strand with the scaffold was found to enhance folding yield. Our results obtained with the 42-helix bundle designed according to rule 3 and 4 support this finding.
A measure for design quality
As the concentration requirements for successful assembly in sodium chloride did not only depend on global shape alone but also on internal design details, we searched for a design-specific quantity that correlated with the required sodium chloride concentration. We analysed all objects tested here regarding the sequence-specific thermal stability of continuous dsDNA domains defined in the design. To this end, we assumed isolated domains, two-state hybridization and 20 mM magnesium in solution (Methods). We found that the percentage of staple DNA strands within each object that by design do not form at least one dsDNA domain with the scaffold with a melting temperature greater than 45 °C correlated strongly with the minimally required sodium concentrations (Fig. 2c). In the six-helix bundle, a minority of staple strands (22%) lacked such stable domains, whereas in the 100-helix bundle it was the majority (79%). The six-helix bundle assembled in the presence of 200 mM sodium chloride, whereas the 100-helix bundle did not form at all, even in the presence of up to 3.4 M sodium chloride. The 42-helix bundle version (rule 4) that required the least sodium chloride (1 M) and gave best folding results had the lowest fraction of staple strands lacking at least one stable dsDNA domain (39%), whereas the version that gave worst results and required the most sodium chloride had the highest content of 'unstable' staple strands (69%).
These findings suggested that minimizing the fraction of staple strands that do not form stable dsDNA domains with the scaffold might be a design strategy that supports fast and high-yield assembly of multi-layer DNA objects in the presence of sodium and that may also positively affect the yield of assembly in the presence of magnesium. Distributing backbone nicks in different ways as tested here is one option to rationally affect the fraction of 'unstable' dsDNA domains, the choice of scaffold sequence may be another that remains to be explored.
Two of the backbone nick distribution rules that we tested (rules 3 and 4) resulted in a similar overall content of long (≥14) dsDNA domains formed between staples and scaffold, but different percentages of staple strands lacking stable dsDNA domains formed with the scaffold (51% and 39%, respectively). Rule 3 considered half-crossovers to achieve at least one 14-bp-long fragment per staple strand. In magnesium chloride reactions, both objects gave comparable results (Fig. 3c), but in sodium chloride reactions the rule 4 object performed significantly better.
Discussion
To conclude, we reported conditions for assembling multi-layer DNA objects in the presence of monovalent sodium ions. Computing the percentage of staple strands in an object that do not form stable dsDNA domains with the scaffold (Tm>45 °C at 20 mM magnesium) and using our tabulated data in Fig. 2c may help estimating the required concentrations for folding other objects in the presence of sodium chloride. For the convenience of the interested reader, we have included as a supplement a graphical user interface (GUI)-based software tool that analyses DNA origami design files in this regard. Our results (and that of Shih and coworkers34 for the case of folding in magnesium) show that minimizing the percentage of weakly binding staple strands through decisions taken during design can positively affect the yield of self-assembly, both in the presence of either sodium or magnesium. We believe that considering the sequence-specific thermal stability of dsDNA domains formed throughout the target object may be a viable starting point towards development of quantitative design approaches for creating 'super-folding' functional DNA objects.
Methods
Molecular self-assembly with DNA origami
Structures were designed using caDNAno17, see Supplementary Fig. S5 for designs. Recombinant M13 filamentous bacteriophage DNA were prepared as previously described21. Staple oligonucleotide strands were synthesized and purified by Eurofins MWG Operon (Ebersberg, Germany, HPSF purification). Production of DNA objects was performed in one-pot reactions by mixing 10 nM scaffold strands derived from M13 bacteriophage with 100 nM of each oligonucleotide staple strand in a pH=8 buffer created by mixing 5 mM TRIS-Base with 1 mM EDTA. Buffer pH was not titrated to avoid the further addition of ionic species. A specified concentration of either magnesium chloride or sodium chloride was created. The mixtures were subjected to thermal-annealing ramps that cooled from 65 to 25°C over the course of minimum 1.5 or up to 23 days (see Supplementary Tables S1–S6). The required minimum salt concentrations for nanostructure formation were determined electrophoretically as described in the results section, using gel image data from the longest annealing protocols where we could still discern bands in ethidium-bromide-stained agarose gels.
Gel electrophoresis
Assembly reaction products were electrophoresed on 2% agarose gels containing 0.5× TBE, 11 mM magnesium chloride, ~0.5 μg ml−1 ethidium bromide at 70 V in a gel box immersed in an ice water bath.
TEM and image processing
Purified particles or unpurified reaction products were adsorbed on glow-discharged formvar-supported carbon-coated Cu400 TEM grids (Science Services, Munich, Germany) and stained using a 2% aqueous uranyl formate solution containing 25 mM sodium hydroxide. Imaging was performed using a Philips CM100 electron microscope operated at 100 kV. Images were acquired using an AMT 4 Megapixel CCD camera. Micrograph scale bars were calibrated by imaging 2D catalase crystals and using the lattice constants as length reference. Imaging was performed at X11 500 and X28 500 magnification. For image processing, libraries of indiviual particle micrographs were created by particle picking using the EMAN2 (ref. 35) boxing routine. Unsupervised particle classification and generation of class averages was performed using Xmipp36.
Design analysis
We developed a custom computer code for parsing caDNAno17 nanostructure design files (Supplementary Software) as a GUI-based software tool (design_analyzer.app) that runs under the National Instruments LabView Runtime for Mac OS X. The code identifies all continuous double-helical DNA domains in a given caDNAno design file, and determines their sequences based on a defined input scaffold sequence permutation. For Fig. 2c, the free energy for DNA hybridization at a given temperature was computed assuming strands at 10 nM and 100 nM concentration for each double-helical DNA domain and 20 mM magnesium chloride in solution. Nearest-neighbor free-energy parameters at 1 M sodium chloride were taken from SantaLucia et al.37 and a salt correction was employed38. The melting point Tm=dH/dS was determined and tabulated for each double-helical DNA domain formed by each staple DNA strand of the nanostructure. The staple strands that do not form at least one double-helical DNA domain with Tm >45 °C were counted and divided by the total number of staple strands in the object.
Additional information
How to cite this article: Martin, TG and Dietz, H. Magnesium-free self-assembly of multi-layer DNA objects. Nat. Commun. 3:1103 doi: 10.1038/ncomms2095 (2012).
References
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1983).
Seeman, N. C. Nanomaterials based on DNA. Annu. Rev. Biochem. 79, 65–87 (2010).
Winfree, E., Furong, L., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Yurke, B., Turberfield, A. J., Mills, A. P., Simmel, F. C. & Neumann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608.
Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA Brownian motor with coordinated legs. Science 324, 67–71 (2009).
Zhang, C. et al. DNA-directed three-dimensional protein organization. Angew. Chem. Int. Ed. 51, 3382–3385 (2012).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Woo, S. & Rothemund, P. W. K. Programmable molecular recognition based on the geometry of DNA objects. Nat. Chem. 3, 620–627 (2011).
Endo, M., Sugita, T., Katsuda, Y., Hidaka, K. & Sugiyama, H. Programmed-assembly system using DNA jigsaw pieces. Chem. Eur. J. 16, 5362–5368 (2010).
Ke, Y. et al. Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano Lett. 9, 2445–2447 (2009).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
Kuzuya, A. & Komiyama, M. Design and construction of a box-shaped 3D-DNA origami. Chem. Commun. 28, 4182–4184 (2009).
Han, D. et al. DNA origami with complex curvature in three-dimensional space. Science 332, 342–346 (2011).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Ke, Y. et al. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 131, 15903–15908 (2009).
Ke, Y., Voigt, N. V., Gothelf, K. & Shih, W. M. Multilayer DNA origami packed on hexagonal and hybrid lattices. J. Am. Chem. Soc. 134, 1770–1774 (2012).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Douglas, S. M., Chou, J. J. & Shih, W. M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA 104, 6644–6648 (2007).
Berardi, M. J., Shih, W. M., Harrison, S. C. & Chou, J. J. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature 476, 109–113 (2011).
Kershner, R. J. et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 4, 557–561 (2009).
Maune, H. T. et al. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat. Nanotechnol. 5, 61–66 (2010).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic objects with tailored optical response. Nature 14, 311–314 (2012).
Wei, R., Martin, T. G., Rant, U. & Dietz, H. DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. 51, 4864–4867 (2012).
Sharma, J. et al. Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science 323, 112–116 (2009).
Duckett, D. R., Murchie, A. I. H. & Lilley, D. M. J. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 9, 583–590 (1990).
Yang, W., Lee, J. Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006).
Kring, J. P. & Williams, J. N. Interference in the fluorometric analysis of pyridine nucleotides by certain ions. J. Biol. Chem. 212, 751–755 (1955).
Sharma, J., Chhabra, R., Liu, Y., Ke, Y. & Yan, H. DNA-templated self-assembly of two-dimensional and periodical gold nanoparticle arrays. Angew. Chem. Int. Ed. 45, 730–735 (2006).
Carter, J. D. & LaBean, T. H. Nanomaterials via programmable DNA self-assembly and peptide molecular recognition. ACS Nano 5, 2200–2005 (2011).
Ke, Y., Bellot, G., Voigt, N. V., Fradkov, E. & Shih, W. M. Two design strategies for enhancement of multilayer–DNA-origami folding: underwinding for specific intercalator rescue and staple-break positioning. Chem. Sci. 3, 2587–2597 (2012).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Scheres, S. H. W. et al. Image processing for electron microscopy single-particle analysis using Xmipp. Nat. Protocols 3, 977–990 (2008).
SantaLucia, J. & Hicks, D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 33, 415–440 (2004).
Owczarzy, R., Moreira, B. G., You, Y., Behlke, M. A. & Walder, J. A. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry 13, 5336–5353 (2008).
Acknowledgements
We thank Christian Wachauf for designing the 6-, 8-, 10- and 12-helix bundles, Fabian Kilchherr for designing the 18-helix bundle, Carlos Castro for designing the 24-helix bundle, and Felix Ritort, Robijn Bruinsma, Thomas Gerling and Jean-Philippe Sobczak for discussions. This work was supported by an European Research Council Starting Grant (GA # 256270) to H.D. Further financial support by the DFG through the Center for Integrated Protein Science Munich (CIPSM) and from the Institute for Advanced Study of the Technische Universität München is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
T.M. performed research, H.D. designed research. T.M. and H.D. analysed data, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures and Tables
Supplementary Figures S1-S5 and Supplementary Tables S1-S6 (PDF 26488 kb)
Supplementary Software
The software, design_analyzer.app, is a Mac OS X application and requires Labview 9.0 runtime for Mac OS X. (ZIP 1429 kb)
Supplementary Data 1
DNA sequences for all objects (XLS 274 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Martin, T., Dietz, H. Magnesium-free self-assembly of multi-layer DNA objects. Nat Commun 3, 1103 (2012). https://doi.org/10.1038/ncomms2095
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms2095
This article is cited by
-
Shaken, not heated: DNA self-assembly at room temperature
Nature Nanotechnology (2023)
-
A temporally resolved DNA framework state machine in living cells
Nature Machine Intelligence (2023)
-
Suppressing high-dimensional crystallographic defects for ultra-scaled DNA arrays
Nature Communications (2022)
-
DNA nanostructures as templates for biomineralization
Nature Reviews Chemistry (2021)
-
DNA origami
Nature Reviews Methods Primers (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.