Abstract
Mycobacterium tuberculosis remains a global threat to human health, yet the molecular mechanisms regulating immunity remain poorly understood. Cytokines can promote or inhibit mycobacterial survival inside macrophages and the underlying mechanisms represent potential targets for host-directed therapies. Here we show that cytokine-STAT signalling promotes mycobacterial survival within macrophages by deregulating lipid droplets via ATG2 repression. In Drosophila infected with Mycobacterium marinum, mycobacterium-induced STAT activity triggered by unpaired-family cytokines reduces Atg2 expression, permitting deregulation of lipid droplets. Increased Atg2 expression or reduced macrophage triglyceride biosynthesis, normalizes lipid deposition in infected phagocytes and reduces numbers of viable intracellular mycobacteria. In human macrophages, addition of IL-6 promotes mycobacterial survival and BCG-induced lipid accumulation by a similar, but probably not identical, mechanism. Our results reveal Atg2 regulation as a mechanism by which cytokines can control lipid droplet homeostasis and consequently resistance to mycobacterial infection in Drosophila.
Similar content being viewed by others
Introduction
Virulent mycobacteria are critically dependent on their ability to survive within macrophages to cause disease. The tendency of macrophages to support or kill mycobacteria is directly regulated by distinct cytokine–JAK–STAT pathways: for example, the Th1 cytokine interferon-γimproves the ability of the macrophage to kill mycobacteria, whereas interleukin (IL)-10 and the Th2-derived cytokines IL-4 and IL-13 can inhibit killing of intracellular mycobacteria1,2,3. Our knowledge of the in vivo functions of these signals in mycobacterial infection, and of their transcriptional targets and the associated physiologies, remains incomplete, especially for those signals that inhibit killing of intracellular bacteria4,5. The genetic and cell-biological mechanisms that underlie these responses are potentially fertile ground for host-targeted therapies for tuberculosis6,7.
The fruitfly Drosophila melanogaster is a well-established model for the study of innate immunity8. Many aspects of the innate immune response to microbes are conserved between flies and vertebrates, including the central role of nuclear factor-κB family transcription factors and Toll-like receptors, anti-inflammatory effects of transforming growth factor-βfamily signals and the presence of bactericidal phagocytes homologous to vertebrate macrophages9,10,11. We have developed a system in which Drosophila are infected with Mycobacterium marinum, which allows us to identify host mechanisms involved in mycobacterial infection12,13. As in mammals, cytokine production by phagocytes is a critical component of the cellular response to infection. Flies have three known genes encoding IL-like signals, upd1, upd2 and upd3 (ref. 14). These proteins have structural and functional similarity to the four-helix-bundle cytokines of mammals15. They bind a GP130-like receptor, encoded by domeless, to activate a conserved JAK–STAT signalling cassette16. Upon bacterial infection in adult flies, upd3 is expressed by phagocytes and activates the JAK–STAT pathway in the fat body, where it is required for infection-induced expression of the stress peptide TotA14. This signalling pathway is critically required for antiviral defense17, it regulates larval haematopoiesis18 and it is important in maintaining gut integrity in response to bacterial infection and other stresses19,20,21,22,23. Other roles in immune defense against bacteria are unknown.
Here we show that unpaired-family cytokines are critical regulators of defense against intracellular virulent mycobacteria. We show that M. marinum infection drives production of upd3 in phagocytes, and that reception of this signal by phagocytes is detrimental to the host. JAK–STAT blockade increased resistance to infection: it prolonged survival of the host, reduced mycobacterial numbers and delayed immune cell death, with similar effects observed in vitro. This effect was associated with overexpression of the autophagy-related gene Atg2 and not other autophagy genes, in vivo and in vitro. M. marinum infection of cultured phagocytes increased intracellular neutral lipids and drove accumulation of unusually large lipid droplets (LDs). Atg2 overexpression or JAK–STAT inhibition reduced intracellular mycobacterial number and partially normalized phagocyte lipid droplet size without significantly changing bulk autophagy. Direct inhibition of triglyceride synthesis prevented the infection-induced alteration in LDs, reduced mycobacteria–LD association and reduced intracellular viable mycobacteria. We then tested these findings in human cells. Similar to Drosophila, IL-6 signalling in macrophages increased intracellular Bacillus Calmette–Guérin (BCG) and Mycobacterium tuberculosis numbers in a dose-dependent manner; ATG2A expression was also reduced, although only to a very small extent. Importantly, the ability of IL-6 to increase the number of viable intracellular mycobacteria was abolished by inhibition of macrophage triglyceride synthesis. Together, these data indicate that loss of a STAT-activating cytokine pathway can reduce survival of intracellular mycobacteria via effects on cellular lipid deposition. In Drosophila, one responsible effector may be ATG2.
Results
unpaired signalling decreases resistance to M. marinum
The IL-6-like cytokine upd3 can be induced in haemocytes by infection with non-pathogenic bacteria; however, the ability of bacterial pathogens to induce its expression has not been previously explored14. This was particularly in question with M. marinum, which is not a strong agonist of known pattern-recognition pathways in Drosophila12. We thus examined the in vivo expression of upd3 during the initial phase of M. marinum infection using an upd3 green fluorescent protein (GFP) reporter and a haemocyte-specific dsRed nuclear marker10,14. We observed strong GFP induction in adult Drosophila haemocytes after mycobacterial infection (Fig. 1a), indicating that mycobacterial infection was indeed sufficient to drive upd3 production from macrophages in vivo. This effect was also visible by quantitative reverse transcriptase–PCR (qRT–PCR) in samples from whole flies (Fig. 1b).
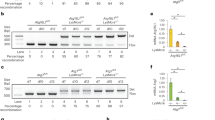
(a) upd3 expression in haemocytes after M. marinum infection. Left, intravital imaging of eGFP expressed under the control of upd3-Gal4 in flies injected with PBS or M. marinum (5,000 CFU). Haemocytes are labelled with nuclear dsRed directly driven by the HmlΔ promoter10. This image was taken 48 h after injection. Scale bar, 10 μm. Right, quantification of the fraction of GFP-positive cells from seven flies based on images similar to the one shown. Error bars show s.d. Values are statistically different (***P<0.001) by Mann–Whitney test. (b) mRNA expression of upd1 and upd3 in wild-type (w1118) and oss flies, uninfected or 72 h after infection with M. marinum. Assayed by qRT–PCR and normalized to Rpl1 (as a loading control), and then to uninfected wild-type flies for each gene. n=4 for each condition. Data were log-transformed before statistical comparison by Student’s t-test (***P<0.001). (c) M. marinum burden in wild-type and oss mutant flies, assayed by qRT–PCR. Flies were injected with 500 CFU M. marinum and kept at 25 °C. P-value for differences in bacterial numbers by two-way analysis of variance (ANOVA), n=6 per time point except for day 9 where n=3 for oss flies. (d) Survival of wild-type and oss-mutant flies, injected with PBS or infected with 500 CFUs M. marinum, as indicated. Flies were kept at 25 °C. P-value for difference between infected survival curves by log-rank test. (e) M. marinum burden in control (w1118;; crq-Gal4/+) and haemocyte-specific upd3-knockdown (w1118; UAS-upd3-IR/+; crq-Gal4/+) flies, assayed by qRT–PCR. Flies were injected with 500 CFU M. marinum and kept at 25 °C. Here and elsewhere, ‘crq>upd3-IR’ is used as an abbreviation for ‘crq-Gal4, UAS-upd3-IR’. P-value for differences in bacterial numbers by one-way ANOVA, n=3 per time point (*P<0.05). (f) Survival of control (w1118;; crq-Gal4/+) and haemocyte-specific upd3-knockdown (w1118; UAS-upd3-IR/+; crq-Gal4/+) flies, injected with PBS or 500 CFU M. marinum. Flies were kept at 25 °C. P-value for difference between infected survival curves by log-rank test.
To test the role of upd3 in mycobacterial infection in Drosophila, we infected flies carrying the oss mutation with M. marinum. oss affects expression of the cytokines upd1 and upd3, and impairs induction of upd3 by infection with non-pathogenic bacteria14,24. We observed that oss mutants did not activate upd3 expression to the same degree as wild-type flies after M marinum infection, whereas upd1 expression was not induced by M. marinum infection in wild-type flies or oss mutants (Fig. 1b). Despite identical starting inocula, oss mutants carried a much lower mycobacterial burden at late stages of infection and survived longer than wild-type controls (Fig. 1c,d).
Although haemocytes were the only visible site of upd3 induction in M. marinum-infected upd3>GFP flies, it remained possible that some other tissue was contributing to the relevant UPD3 pool. Moreover, although we saw no significant induction of upd1 by infection, changes in UPD1 might still contribute to the oss phenotype. To verify that haemocyte-derived UPD3 was responsible for the mycobacterial resistance phenotype of oss mutants, we knocked down upd3 with the haemocyte-specific crq-Gal4 driver and examined mycobacterial pathogenesis25. Haemocyte upd3 knockdown gave strong lifespan extension after infection and increased mycobacterial number (Fig. 1e,f). Together, these experiments indicate that haemocytes produce upd3 upon mycobacterial infection and haemocyte-derived upd3 shortens the infected lifespan of the host and impairs resistance against M. marinum.
Haemocyte STAT activity decreases resistance to mycobacteria
To identify relevant UPD3 target tissues, we used RNA interference (RNAi) knockdown of Stat92E (the sole Drosophila STAT). Prior work has suggested that immune-induced STAT signalling is important in regulating humoral immune responses via effects on the Drosophila fat body26. However, Stat92E knockdown in the fat body had no effect on survival after M. marinum infection (Supplementary Fig. 1a). The primary direct immune effect of the fat body is the immune-inducible expression of antimicrobial peptides; we found that oss mutant flies exhibited no consistent differences in antimicrobial peptide expression before haemocyte death (Supplementary Fig. 1b–e). We thus concluded that the effect of upd3 on mycobacterial infection was independent of the fat body.
We next tested the effect of cytokine signalling on haemocytes themselves. As the JAK–STAT pathway regulates larval haematopoiesis, we combined our haemocyte drivers with Gal80ts, making their effect temperature sensitive, to enable adult-specific Stat92E knockdown18,27,28. Adult-specific Stat92E knockdown or expression of a dominant-negative form of the upd3 receptor dome in haemocytes significantly improved resistance to infection (Fig. 2a,b and Supplementary Fig. 2). These results together indicate that haemocyte-derived upd3 activates STAT signalling in haemocytes, decreasing resistance to mycobacterial infection and impairing host survival.
(a) M. marinum burden in control (w1118; tub-Gal80ts/+; crq-Gal4/+) and inducible haemocyte-specific Stat92E-knockdown (w1118, UAS-Stat92E-IR/+; UAS-Stat92E-IR/tub-Gal80ts; crq-Gal4/+) flies, assayed by qRT–PCR. Flies were initially infected with 500 CFU M. marinum and cultured at 29 °C. P-value for difference in bacterial numbers by two-way analysis of variance (ANOVA), n=5 per timepoint. (b) Survival of control (w1118; tub-Gal80ts/+; crq-Gal4/+) and haemocyte-specific Stat92E-knockdown (w1118, UAS-Stat92E -IR/+; UAS-Stat92E -IR/tub-Gal80ts; crq-Gal4/+) flies, injected with PBS or infected with 500 CFU M. marinum, as indicated. Infectious dose and temperature as in a. P-value for difference between infected survival curves by log-rank test. (c) Fold change in expression of several autophagy genes in Stat92E-IR (w1118, UAS-Stat92E-IR; UAS-Stat92E-IR/tub-Gal80ts; crq-Gal4/+) flies compared with controls (w1118; tub-Gal80ts/+; crq-Gal4/+and w1118, UAS-Stat92E-IR; UAS-Stat92-IR/+) 5 days after injection with 500 CFU M. marinum. The value shown is the geometric mean of two ratios (knockdown/driver-only control and knockdown/STAT-IR only control). Atg2 fold change is statistically different from other autophagy genes (***P<0.001 by Mann–Whitney test, n=14). (d) Atg2 expression by qRT–PCR in S2R+ cells overexpressing HA (control) or upd3. Normalized to Rpl1. Values are statistically different (*P<0.05 by Mann–Whitney test, n=6). (e) Expression of Atg2 by qRT–PCR in S2R+ cells with Luciferase or Stat92E knocked down, with or without M. marinum infection. Normalized to Rpl1 and then to the uninfected Luciferase control. Values are statistically different as indicated (**P<0.01 by Mann–Whitney test, n=7). (f) Intracellular dsRed-expressing M. marinum in S2R+ cells overexpressing HA (control) or Atg2. Picture is a representative; graph shows normalized M. marinum fluorescence from 37 (Atg2 O/E) or 45 (control) cells. Values are statistically different (*P<0.05 by Mann–Whitney test). Scale bar, 5 μm. (g) Intracellular dsRed-expressing M. marinum in S2R+ cells with Luciferase or Stat92E knocked down by RNAi. The graph shows normalized M. marinum fluorescence from 44 (Stat dsRNA) or 76 (Luc dsRNA) cells. Values are statistically different (**P<0.01 by Mann–Whitney test).
Cell death via necrosis or apoptosis is commonly observed in vertebrate macrophages infected with M. tuberculosis or M. marinum29,30,31. We observed marked cytotoxicity due to M. marinum infection in Drosophila S2R+ cells, an embryonic-derived macrophage cell line, and progressive loss of haemocytes during the last few days of M. marinum infection in vivo (Supplementary Fig. 3a–d). Just before dying, flies harboured no visible haemocytes and dramatically reduced expression of the haemocyte marker Hml was detected by qRT–PCR on the whole fly (Supplementary Fig. 3c–e). The oss mutation improved haemocyte survival in infected flies (Supplementary Fig. 3b–d). Importantly, this was not a result of differential microbial internalization, as the fraction of haemocytes containing M. marinum was not different between control and upd3-IR flies 16 h after infection (Supplementary Fig. 4a,b). The difference in haemocyte survival, such as the difference in haemocyte number, was also seen in flies with adult-specific haemocyte Stat92E knockdown (Supplementary Fig. 4c–e). We thus conclude that upd3-STAT signalling alters the interaction between mycobacteria and phagocytes so as to impair the survival of infected cells.
JAK–STAT activation in haemocytes downregulates Atg2
We wished to identify target genes and cell-biological processes behind the reduced mycobacterial numbers seen as a result of impaired cytokine signalling. Activation of JAK–STAT signalling in Kc167 cells reduces expression of several autophagy-related genes32. As phagocyte autophagy can impair intracellular survival of M. tuberculosis33, we tested this transcriptional effect in vivo, analysing the expression levels of 14 autophagy-related genes in flies with haemocyte-specific Stat92E knockdown. Surprisingly, Atg2 was the only autophagy gene with strongly enhanced expression following mycobacterial infection in Stat92E knockdown flies (Fig. 2c). Further analysis revealed that Atg2 overexpression was seen in haemocyte-specific Stat92E and upd3 knockdown flies independent of infection (Supplementary Fig. 5a,b). Conversely, in vitro, upd3 overexpression induced the known JAK–STAT pathway target TotA and reduced Atg2 expression (Fig. 2d and Supplementary Fig. 5c,d). The specific effect on Atg2 expression was particularly interesting, as in HeLa cells Atg2 is separably important for autophagy and neutral lipid distribution, both processes associated with intracellular survival or killing of mycobacteria33,34,35,36.
To clarify how cytokines interacted with other inflammatory cues to regulate phagocyte Atg2, we measured expression of Atg2 in S2R+ cells with or without Stat92E knockdown and with or without M. marinum infection. Stat92E knockdown or M. marinum infection gave mild or no increase in Atg2 expression but, when combined, these two stimuli strongly induced Atg2 expression, suggesting that upd3 limits infection-induced Atg2 overexpression (Fig. 2e). The difference observed between the in vivo context (where infection is not required for increased Atg2 expression in Stat92E knockdowns) and these in vitro observations may be the result of the different cytokine milieu experienced by cells in culture, compared with those in the animal, or to the contribution of other tissues.
Increased Atg2 impairs intracellular M. marinum survival
As Atg2 was specifically affected by loss of upd3-Stat92E signalling, we assayed the effect of Atg2 overexpression in cultured phagocytes. Overexpression of GFP-ATG2 or RNAi of Stat92E reduced by 50% the number of intracellular mycobacteria per cell and eliminated mycobacterium-induced phagocyte death (Fig. 2f,g and Supplementary Fig. 5e,f). The effect of Atg2 on resistance to infection was not due to general changes in particle internalization, as Atg2 overexpression did not impair uptake of pHrodo-labelled beads (Supplementary Fig. 5g).
Atg2 knockdown has separable effects on autophagy and lipid deposition in HeLa cells35. We assayed the effect of Atg2 overexpression on autophagy and found no clear differences in autophagosome number, processing of ATG8 or association of autophagosomes with mycobacteria (Supplementary Fig. 6).
M. marinum benefits from neutral lipids in phagocytes
In M. tuberculosis infection, some macrophages accumulate LDs, which has been suggested to promote mycobacterial persistence within the host37,38. Indeed, intracellular survival of M. tuberculosis depends on its ability to recalibrate macrophage lipid homeostasis34,39. M. marinum infection, similar to M. tuberculosis infection, induces lipid accumulation in Drosophila cells (Fig. 3a). As we had seen little effect on general autophagy from Atg2 overexpression, we explored the interaction of Atg2 with LDs. GFP-ATG2 associated with many LDs, as previously seen in HeLa cells, and sometimes appeared to coat intracellular mycobacteria (Fig. 3b and Supplementary Fig. 6a)35.
(a) Neutral lipids (stained with Oil Red O) and intracellular mycobacteria (stained with fluorescein isothiocyanate-anti-M tuberculosis) in cells variously uninfected, infected with live M. marinum or infected with heat-killed (H/K) M. marinum. (b) Co-localization of ATG2-GFP with LipidTox-stained intracellular neutral lipids. Cells were observed 48 h after transfection. Scale bars, 5 μm. (c) Quantification of neutral lipid volume per cell and total number of LDs with at least one axis >1 μm in length in cells treated as in b. Values are statistically different (**P<0.01 and ***P<0.001) by Mann–Whitney test. n=130 for non-infected cells, n=110 for M.m.-infected cells and n=96 for cells in contact with heat-killed M.m. (d) Number of LDs (stained with BODIPY 500/510) of volume ≥1.5 μm3 in cells treated with dsRNA for mdy or Luc, and infected with dsRed M. marinum. Values are statistically different (***P<0.001 by Mann–Whitney test, n=71 for mdy dsRNA, n=135 for Luc dsRNA). (e) Number of LDs (stained with BODIPY 500/510) at a distance ≤1 μm from bacteria and measured per unit of bacterial volume. Cells were treated with dsRNA for mdy or Luc and infected with dsRed M. marinum. Values are statistically different (***P<0.001 by Mann–Whitney test, n=64 for mdy dsRNA, n=119 for Luc dsRNA). (f) Co-localization of BODIPY-stained LDs with dsRed M. marinum in S2R+ cells with or without mdy knockdown. Scale bar, 5 μm. (g) Quantification of bacterial volume per cells in S2R+ cells treated with dsRNA for mdy or Luc and infected with dsRed M. marinum. Values are statistically different (***P<0.001 by Mann–Whitney test, n=64 for mdy dsRNA, n=119 for Luc dsRNA).
Quantitative analysis of lipid deposition in infected cells revealed a mild increase in total intracellular neutral lipid, but a dramatic increase in the number of unusually large LDs in infected samples (Fig. 3c). Heat-killed M. marinum induced greater accumulation of LDs and more large lipid structures than live bacteria (Fig. 3c), whereas infection of S2R+ cells with Escherichia coli, as an activator of general inflammatory signals, did not alter neutral lipids (Supplementary Fig. 8a,b). Although we were able to stain neutral lipids within free mycobacteria, the size and shape of the lipid inclusions we observed in infected phagocytes were incompatible with the lipids being exclusively intrabacterial and the large irregular lipid inclusions we observed were often but not always physically associated with intracellular mycobacteria as detected by immunofluorescence or fluorescent protein expression (Fig. 3a,b and Supplementary Fig. 8c–e).
The fact that dead mycobacteria drove lipid accumulation and that large lipid inclusions were only partially associated with live intracellular M. marinum implied that these aberrant large lipid inclusions were host-derived and resulted from some change in host cell lipid deposition or metabolism exploited by mycobacteria. We tested this directly by RNAi inactivation of midway (mdy), the Drosophila homologue of diacylglycerol O-acyltransferase 1 (DGAT1), the final enzyme in triglyceride synthesis40. mdy knockdown reduced LD numbers and volume in uninfected and M. marinum-infected S2R+ cells (Supplementary Fig. 9a–f). We identified a threshold LD volume associated with pathogenesis by determining the cumulative distribution function of LD volumes in uninfected and M marinum-infected cells. These functions intersected at a volume of 1.5 μm3, indicating that most droplets of volume greater than this in infected cells were due to mycobacterial activity (Supplementary Fig. 7g). mdy knockdown impaired M. marinum-induced formation of large LDs and reduced mycobacterial association with LDs, leading to a general reduction in bacterial burden per cell (Fig. 3d–g). These data demonstrate that M. marinum survival and/or proliferation is dependent on lipid accumulation in Drosophila phagocytes.
Increased Atg2 expression reduces large LDs
As preventing LD accumulation during infection reduced mycobacterial numbers and as ATG2 associated with LDs and reduced bacterial load, we hypothesized that ATG2 affected resistance to M. marinum by altering lipid deposition in immune cells. We tested this by infecting Atg2-transfected and control cells with M. marinum and observing LD volume (Fig. 4a–c). M. marinum-infected cells overexpressing Atg2 exhibited a marked reduction in the number of large LDs compared with infected controls. Stat92E knockdown, which induced Atg2 expression (Fig. 2e), also decreased large LD size in infected cells (Fig. 4d). Stat92E knockdown did not affect LD size in uninfected cells, in keeping with our observation that this knockdown did not affect Atg2 expression in the absence of infection (Fig. 2e). These data together suggest that overexpression of Atg2 observed in upd3/Stat92E knockdown flies renders phagocytes inhospitable to mycobacteria via effects on lipid deposition.
(a) Intracellular neutral lipid in M. marinum-infected cells overexpressing either HA (control) or Atg2. Scale bars, 5 μm. (b) Quantification of neutral lipid volume per cell and total number of LDs with at least one axis >1 μm in length in cells treated as in a. A slight increase in the overall volume of neutral lipids was observed in Atg2-overexpressing cells compared with control (*P<0.05 and **P<0.01, Mann–Whitney test, n=130 for non-infected controls, n=110 for Mm-infected controls, n=107 for non-infected cells overexpressing Atg2 and n=100 for infected cells overexpressing Atg2). (c) Distribution of large (>1.5 μm3) LD volumes in cells overexpressing Atg2 or HA and with the infections indicated. Volumes <1.5 μm3 are not shown but were included in statistical analysis. Punctae (3,100–5,000) were analysed depending on genotype and condition. Significance values are from the Kolmogorov–Smirnov test. (d) Distribution of large (>1.5 μm3) LD volumes in cells with Luciferase or Stat92E knocked down by RNAi, with or without infection as indicated. Volumes <1.5 μm3 are not shown but were included in statistical analysis. Punctae (1,100 to 2,000) were analysed depending on genotype and condition. Significance values are from the Kolmogorov–Smirnov test.
IL-6 impairs mycobacterial killing via effects on triglyceride
To see whether our results were relevant to human mycobacterial disease, we tested the ability of several JAK–STAT activating cytokines to impair killing of Mycobacterium bovis BCG by THP-1 cells. IL-6, added 2 h after infection, strongly inhibited the mycobactericidal activity of these cells (Fig. 5a). To further clarify the timing of this effect, we measured viable intracellular BCG in cells treated before and/or after infection with IL-6; we observed that either pre-, post- or pre- and postinfection IL-6 caused a significant increase in viable intracellular BCG at 24 and 48 h (Fig. 5b). We also observed that IL-6 caused a dose-dependent increase in survival of an auxotrophic mutant of M. tuberculosis within primary human macrophages (Fig. 5c). We then tested the effect of ATG2A overexpression on intracellular lipid stores in THP-1 cells and observed that overexpression of ATG2A was sufficient to markedly reduce neutral lipid stores in these cells (Fig. 5d). upd3 has been suggested to be an evolutionary homologue of IL-6, and IL-6 is inducible by M. tuberculosis and can prevent killing of intracellular mycobacteria15,41,42; this suggested that the mechanisms of IL-6 action on human macrophages might be similar to the effects of upd3 in Drosophila.
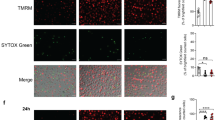
(a,b) Screening of the effect of cytokines signaling through JAK–STAT pathway on intracellular survival of mycobacteria in human macrophages. (a) Differentiated THP-1 cells were incubated with a luminescent strain of M. bovis BCG (BCG-lux) for 2 h, repeatedly washed, then treated with cytokines (20 ng ml−1) for 24 h at 37 °C. Cells were then lysed and luminescence measured. Viable intracellular mycobacteria were increased by IL-6 (red) and IL-5, and reduced by granuloyte–macrophage colony-stimulating factor and Leptin (green). Significance comparisons are between indicated treatment and untreated control. (b) Comparison of effects of treatment with IL-6 before and after infection in THP-1 cells. (c) Primary human macrophages were incubated with Bleupan M. tuberculosis for 2 h, repeatedly washed and then treated with IL-6 (red, at concentrations shown) for 24 h at 37 °C before cells were lysed and plated to count CFUs. Significance comparisons are between indicated treatment and untreated control. (d) ATG2A-V5 overexpression reduces intracellular neutral lipids in THP-1 cells. Scale bars, 10 μm. (e) Treatment of primary human macrophages with IL-6 (80 ng ml−1 for 24 h) causes a small reduction in ATG2A protein abundance. Quantification is from three independent experiments. Error bars represent s.e.m. (f,g) BCG-induced LD accumulation in enhanced by IL-6 treatment. Representative images (f) and fluorescence quantification (g) of intracellular LDs (red), in primary human macrophages 24 h after infection with GFP-labelled M. bovis BCG (green). LDs are increased following infection, further increased in the presence of IL-6 treatment (80 ng ml−1; red) and significantly reduced by pretreatment with the DGAT1 inhibitor T863 (10 μM, 48 h). Scale bars, 10 μm (f). (h) The increase in intracellular viable M. bovis BCG in primary human macrophages treated with IL-6 (red; 80 ng ml−1 for 24 h post infection) is blocked by pretreatment with the DGAT1 inhibitor T863 (as above). *P<0.05, **P<0.005 and ***P<0.001. Figures are representative of at least three independent experiments with at least three replicates per data point.
We next tested the effect of IL-6 treatment on abundance of ATG2A protein. Although IL-6 treatment caused a statistically significant reduction in ATG2A protein abundance in monocyte-derived macrophages (P=0.0384, unpaired Student’s t-test), this effect was extremely small (Fig. 5e). Thus, although the STAT-ATG2 transcriptional circuit may function in similar ways in human and fly macrophages, it is difficult to imagine that the effect of exogenous IL-6 on mycobacterial number is driven by reductions in total quantity of cellular ATG2A.
We then examined the effect of IL-6 on lipid accumulation in infected primary human macrophages. We treated macrophages with IL-6 with or without pharmacological inhibition of DGAT1. IL-6 treatment potentiated the increase in intracellular lipid deposition driven by mycobacterial infection (Fig. 5f,g). DGAT1 inhibition reduced intracellular triglyceride levels, eliminated the ability of mycobacteria to drive lipid accumulation and abolished the ability of IL-6 to promote intracellular mycobacterial survival (Fig. 5f–h), indicating that the ability of IL-6 to increase intracellular mycobacterial numbers depended on its ability to enhance bacterial access to host triglyceride.
Discussion
Here we identify a molecular mechanism that can reduce mycobacterial viability in phagocytes and show how this mechanism is inhibited by cytokine signalling in vivo. We show that haemocyte-derived cytokine signals activate JAK–STAT signalling in haemocytes in the early phases of M. marinum infection. Inhibiting production or response to this signal in adult haemocytes results in enhanced resistance to mycobacterial infection. We then show that flies in which either upd3 or JAK–STAT signalling is impaired in haemocytes exhibit increased expression of Atg2, but not other autophagy genes. Atg2 overexpression is sufficient to reduce mycobacterial survival in cultured Drosophila phagocytes and to inhibit changes in neutral lipid metabolism observed in M. marinum-infected cells. In parallel, we demonstrate that disturbing pathogen-induced changes in triglyceride metabolism directly impairs bacterial survival. Finally, a similar mechanism appears to function in human macrophages in response to IL-6 signalling. Together, these data indicate that some STAT-activating cytokines increase intracellular mycobacterial survival via effects on triglyceride deposition, that ATG2 regulation is a potential effector and that inhibition of macrophage triglyceride accumulation may be a useful therapeutic approach in mycobacterial disease.
Mycobacterium-infected macrophages often show aberrant accumulation of neutral lipids43, but the biological importance of this event is only partly understood. Intracellular mycobacteria depend on host lipids as a source of nutrients37,39, but this is not necessarily the only important role of neutral lipids in promoting mycobacterial survival. The fact that we observe changes in mycobacterial numbers as a result of triglyceride perturbations as little as 24 h after infection implies that accumulation of triglycerides may also directly disrupt phagocyte killing mechanisms. In this context, it may be relevant that neutral lipids have recently been observed to be protective against oxidative stress in the developing Drosophila central nervous system44, suggesting that lipid accumulation in macrophages might protect intracellular bacteria from oxidative killing mechanisms. Nonetheless, this effect will clearly not be universal: in some contexts, LDs contain large amounts of free histones that can exert direct antibacterial properties45. Another possibility is that the effect on bacterial survival is due to changes in production of lipids used as signalling molecules, such as leukotrienes, lipoxins and other eicosanoids46. Although the roles of these molecules in Drosophila immune responses has not been characterized, they play critical roles in regulation of vertebrate responses to mycobacterial infection47 and it is not difficult to imagine that their production would be altered by the effects on triglyceride deposition we document here. In either case, our observations suggest the possibility that fusion of LDs with bacteria-containing vesicles is normally a productive aspect of the host defense that has been co-opted by some intracellular pathogens to promote their own nutrition and/or protect themselves from intracellular killing—although other explanations for these phenomena cannot be excluded.
Although autophagy can be altered by pro- and anti-inflammatory cytokines and has been argued to be important in clearance of intracellular M. tuberculosis3,33, the in vivo relevance of this effect is not clear48. The role of autophagy in M. marinum infection is similarly unclear, though cytosolic M. marinum can be sequestered in double-membraned vesicles in bone-marrow-derived macrophages independently of the classical autophagy pathway proteins ATG5 or LC3, suggesting an unusual autophagy-like mechanism may be involved in the response to this organism49. Our work suggests that the effects of autophagy on infection and sequestration of mycobacteria and the effects of mycobacteria on intracellular lipid inclusions may be fundamentally linked by common underlying molecular mechanisms. Effects ascribed to autophagic mechanisms may, in fact, be due to autophagy components—in the case of our work, ATG2—acting outside of the normal autophagy pathways to alter lipid accumulation. Future work, in particular to delineate the genetic requirements for lipophagy as opposed to general autophagy, will help resolve these effects in infection.
Our data suggest a model in which STAT-driven Atg2 repression deregulates lipid storage upon infection to the advantage of mycobacteria (Fig. 6). Despite potent bactericidal mechanisms, macrophages are often unable to eradicate M. tuberculosis, which then persists in the organs of the infected individual50. Therefore, novel strategies are needed to counteract bacterial latency and permit killing. Our work suggests two strategies to target mycobacterial infection by altering lipid metabolism in macrophages. One such strategy is direct inhibition of DGAT1. The fact that ATG2A overexpression is sufficient to reduce intracellular triglyceride in cultured THP-1 cells, and that Atg2 overexpression is sufficient to reduce mycobacterial survival in fly cells, suggests another such strategy. The strong and specific repression of Atg2 by JAK–STAT signalling, at least in Drosophila, indicates one regulatory mechanism for this gene, but it is clear that even in this comparatively simple system other regulators are critical. Identification of these other regulatory mechanisms may open new avenues for antimycobacterial therapies.
Upon infection, haemocytes secrete upd3, which activates the JAK–STAT pathway by signalling back to haemocytes. Upon activation of JAK–STAT, Atg2 expression is reduced, large and irregular LDs are formed, and the bacteria survive and proliferate within the immune cell. However, if the JAK–STAT pathway is inhibited, Atg2 expression is increased and ATG2 protein is found surrounding LDs and bacteria (red dots). The LDs maintain small and circular structures and the bacteria are less able to survive or replicate.
Methods
Drosophila stocks
The following stocks were used: from the Bloomington Drosophila Stock Center: oss (BDSC 79), Gal80ts (BDSC 7017, 7018, 7019, 7108), c564 (BDSC 6982), HmlΔ-Gal4 (BDSC 30140) and UAS-Stat92E-IR (BDSC 26899); from the Vienna Drosophila Resource Center: UAS-upd3-IR (VDRC 106869) and UAS-Stat92E-IR (VDRC 106980); from Hervé Agaisse (Yale University): UAS-upd3-IR; from Nathalie Franc (The Scripps Research Institute): crq-Gal4; from James Castelli-Gair Hombría (Centro Andaluz de Biología del Desarrollo): UAS-domeΔcyt.
Survival analysis
Mycobacterial and Drosophila culture and infections were carried out substantially as described, using 5- to 9-day-old male flies12. Injected flies were incubated 20 flies to a vial and the number dead was recorded twice a day; all survival experiments were repeated at least three times with qualitatively similar results. When using a tubulin-Gal80ts construct, crosses were kept at 18 °C and flies were collected and switched to 29 °C the day after eclosion; they were then maintained at 29 °C for the duration of the experiment. For experiments not involving tub-Gal80ts, flies were kept at 25 °C at all times.
Intravital imaging
For haemocyte counts, 5–7-day-old flies carrying HmlΔ-Gal4, UAS-2xeGFP or Crq-Gal4, UAS-2xeGFP were injected with 500 colony-forming unit (CFU) M. marinum and imaged at indicated times. Flies were glued to coverslips and imaged immediately using a confocal microscope (SP5, Leica) equipped with an environmental control chamber. The environmental chamber was set to 25 °C or 29 °C when tub-Gal80ts flies were used. Flies that died during the procedure were excluded from the analysis. GFP was excited at 488 nm with an argon laser and 5.63 μm sections were imaged through the coverslip, glue and dorsal cuticle. Flies carrying HmlΔ-dsRed.nls and upd3-Gal4, UAS-GFP were imaged 48 h after injection of 5,000 CFUs using the same procedure with sequential excitation at 488 and 561 nm, and with images taken every 0.69 μm.
Drosophila cell culture and infection
S2R+ cells (DGRC, Bloomington, Indiana) were maintained in M3 medium supplemented with 10% fetal bovine serum and 50 U ml−1 penicillin–streptomycin. S2R+ cells were transfected using Effectene (Qiagen); 24 h later, they were washed and then infected with M. marinum (suspended in M3 medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin) at a multiplicity of infection (MOI) of 10–20. Samples were collected after 24 or 48 h for western blotting, qRT–PCR or imaging.
For overexpression experiments, GFP-tagged human ATG2A (from plasmid pEGFP-C1-hAtg2A, Addgene, deposited by Noboru Mizushima) and Drosophila upd3 (by PCR from Drosophila complementary DNA) were cloned into pPAC-HA, a vector containing the Drosophila Actin 5C promoter (from Nic Tapon (Francis Crick Institute)). In all overexpression experiments, the displayed control is cells transfected in parallel with the empty vector, in which a 3xHA peptide is expressed.
Phagocytosis assays on cultured cells were done as described51.
For RNAi experiments, cells were bathed in double-stranded RNA (dsRNA) targeting Stat92E or, as a control, Renilla luciferase. RNAi constructs were designed using the SnapDragon web tool with no predicted off-targets; template amplicons were produced by PCR from plasmid or Drosophila genomic DNA with the following primers: T7-RLucL, 5′-TAA TAC GAC TCA CTA TAG GGA GAC TGA TCA AGA GCG AAG AGG G-3′; T7-RLucR, 5′-TAA TAC GAC TCA CTA TAG GGA GAC TTT CAC GAA CTC GGT GTT G-3′; T7-Stat92eL, 5′-TAA TAC GAC TCA CTA TAG GGA GAA AGC TGC TTG CCC AAA ACT A-3′; T7-Stat92eR, 5′-TAA TAC GAC TCA CTA TAG GGA GAG TCG ACG ATA AAG GCA GAG C-3′; T7-mdyL, 5′-TAA TAC GAC TCA CTA TAG GGA GAG CGA CTT CTT AAA CTT GCG G-3′; and T7-mdyR, 5′-TAA TAC GAC TCA CTA TAG GGA GAG ATA TGG CCG ATA GGG GAA T-3′. dsRNA was made with the MEGAscript T7 kit (Ambion). For efficient knockdown, 1.2 μg dsRNA was added per well in 48-well plates containing 200,000–500,000 cells according to the DRSC RNAi protocol. After 3 days of dsRNA treatment, M. marinum was added to the dsRNA solution, at an MOI of 20–60 based on the initial number of cells added in the experiment. Cells were lysed in TRIzol 24–48 h post infection.
Flow cytometry of Drosophila haemocytes and S2R+ cells
For cytotoxicity assays in uninfected and infected Drosophila S2R+ cells, the supernatant medium containing non-adherent cells was collected 24 h post infection and transferred directly into a FACS tube. The adherent S2R+ cells of the same sample were detached using 2 ml of ice-cold culture medium and pipetting up and down. The medium was then added to the previously collected supernatant sample. All samples were filtered with a 70 μm cell strainer filter (BD Biosciences) and kept on ice before FACS analysis. Five minutes before analysis, 5 μl of propidium iodide (PI) was added to 500 μl of samples to stain dead cells. Cells were analysed with a FACS Aria II (Becton Dickinson (BD)), using a 100 μm nozzle and the FACS-Diva software. All experiments included the following controls: M. marinum only samples with and without PI staining, and non-infected cells with and without PI.
For phagocytosis assays with phrodo-coated beads in uninfected and infected Drosophila S2R+ cells, supernatant and adherent cells were collected as described above 48 h post transfection. Cells (500,000) were spun down for 3 min at 21 °C, 1,400 r.p.m. and resuspended in fresh medium. pHrodo-coated beads were added at a concentration of 1/100 v/v and incubated with cells for 30 min at 25 °C. Cells were then spun for 3 min at 21 °C, 1,400 r.p.m. to remove free beads. Fresh medium was added and cells were incubated one more hour before analysis by FACS (BD LRS Fortessa). Samples were analysed in the ultraviolet channel (IndoViolet beads) and the Texas red channel (pHrodo). The experiment included the following controls: cells without beads and beads-only control (no pHrodo).
For measurement of M. marinum uptake by haemocytes in vivo, the different genotypes (HmlΔeGFP/+ and HmlΔeGFP>upd3-IR) were injected with M. marinum expressing Tomato fluorescent protein. Sixteen hours post infection 30–40 flies per sample were anaesthetized with CO2. The flies were carefully transferred onto a 70 μm mesh, and smashed and filtered in 10 ml ice-cold 1 × PBS with 2 mM EDTA. The cell suspension was centrifuged at 150 g for 10 min at 4 °C and the supernatant carefully removed. The cell pellet was washed in 10 ml 1xPBS with 2 mM EDTA twice, afterwards the cells were resuspended in 500 μl of 1 × PBS with 2 mM EDTA and transferred to a FACS tube by filtering the solution through a 70 μm filter mesh. Samples were acquired with a BD Fortessa and analysed using the FlowJo analysis software. Four samples per genotype were analysed.
In vitro mycobacterial infection assays
Luminescent reporter strain infection: a validated luminescent reporter strain of M. bovis BCG (BCG-lux) encoding the Vibrio lux AB gene, generated as described52, was used to infect macrophages. Correlation between CFUs and luminescence was confirmed before experiments. Primary monocyte-derived human macrophages, generated (as previously described53) from healthy consented subjects (Regional Ethics approval: REC: 12/WA/0148 and 4/EE/1187) or THP-1 cells (ATCC), were differentiated by treatment with 5 ng ml−1 PMA 48 h before infection, inoculated with BCG-lux (at an MOI of 5:1) for 2 h at 37 °C, repeatedly washed, then incubated for 24 h at 37 °C in the presence of compounds as indicated. Unless otherwise stated, recombinant cytokines (Peprotech) were used at 20 ng ml−1 and added to cells following 2 h incubation with mycobacteria. Where indicated, cells were pretreated with DGAT1 inhibitor T863 (10 μM) for 48 h and then 24 h post infection, IL-6 was treated for 24 h post infection at 80 ng μl−1. Cells were lysed and luminescence measured as described53. Experiments were carried out in sextuplicate. Results are representative of at least three separate experiments.
M. tuberculosis infection: M. tuberculosis ΔleuD ΔpanCD (Bleupan)54 stably transfected with pMSP12::GFP was grown in Middlebrook 7H9 broth (Difco) containing 0.5% glycerol, 0.05% Tween 80, 10% OADC (BD), 0.05 mg ml−1 L-leucine, 0.024 mg ml−1 calcium pantothenate and 0.2 mg ml−1 Hygromycin B in 250 ml sterile disposable Erlenmeyer flasks with mild agitation at 37 °C. Bacterial cells were harvested at mid log phase by centrifugation at 3,000 g for 7 min, pellets were resuspended in 7H9 broth containing leucine and pantothenate supplements as above plus 18% glycerol and frozen at −80 °C until use. Cells were infected at an MOI of 5:1, lysed at times indicated and CFUs enumerated as described55.
Confocal immunofluorescence on human cells
Primary human macrophages were grown on glass coverslips, infected with M. bovis BCG (BCG-lux) and treated as described, rinsed with PBS, fixed with methanol and permeabilized with 0.1% Triton X-100 (Sigma)56 before being stained with Nile red 1:10,000 (dissolved in isopropanol), rinsed with water and then mounted with ProLong Gold antifade containing DAPI (Invitrogen). Images were acquired on a Zeiss LSM880 confocal microscope (Plan-Apochromat × 63/1.40 Oil-immersion lens) and analysed with Zen 2010 (Carl Zeiss), and fluorescence per cell measured by Volocity software.
Antibodies and western blots
For the anti-ATG2A blotting shown in Fig. 5, primary human macrophages were treated with IL-6 80 ng μl−1 for 24 h before being lysed. The primary antibody used was anti-ATG2A pAb (MBL Life Science) at 1:400. Other western blots were performed with anti-Drosophila Atg8 (courtesy Katja Köhler (University of Zürich)57 and G. Juhasz (Eötvös Loránd University)58, both used at 1:200) and anti-α-tubulin (Developmental Studies Hybridoma Bank 12G10, used 1:10,000). Immunofluorescence on fly cells was performed with fluorescein isothiocyanate-labelled anti-M tuberculosis (Invitrogen PA1-28997, used 1:50) and rabbit anti-GFP (Invitrogen A-11122, used 1:100). Immunofluorescence on human cells was performed with anti-V5 from Novus Biologicals (NB 600-381), 1:400.
M. marinum culture
M. marinum strain M and fluorescent derivatives were grown standing at 25° in Middlebrook 7H9 media, supplemented with OADC, Tween-80, glycerol and antibiotics as necessary to maintain fluorescent plasmids12. Single-cell bacterial suspensions were produced as follows. Cultures were pelleted by centrifugation at 4,000 g for 5 min, then resuspended in PBS+0.2% Tween-80. Clumps were then separated by centrifugation at 200 g for 5 min. The supernatant from this centrifugation contained primarily single bacterial cells. M. marinum strains were all strain M carrying various fluorescent proteins under the control of the msp12 promoter59; all were courtesy of Lalita Ramakrishnan and Antonio Pagan (University of Cambridge).
Statistics
Statistical analyses were performed using GraphPad Prism or R. Two-way analysis of variance, Mann–Whitney test and Kolmogorov–Smirnov test were used in this study. Statistical analysis was only performed on experiments with a least four samples per condition. P-values are as follows: *P<0.05, **P<0.01 and ***P<0.001. Unless otherwise noted, error bars correspond to s.e.m.
qRT–PCR analysis
RNA was extracted from adult flies using TRIzol (Invitrogen) according to the manufacturer’s directions, except that samples from which mycobacterial RNA was amplified were first homogenized in a 3:1 mixture of chloroform and methanol60. Mycobacterial quantification was performed using a standard in which known quantities of fly and bacteria were mixed and then treated the same as injected samples. qRT–PCR was performed as described10,61. PCR primers used in this work are listed in Supplementary Table 1.
Data availability
All relevant data are available from the authors, except where precluded by human subject confidentiality.
Additional information
How to cite this article: Péan, C. B. et al. Regulation of phagocyte triglyceride by a STAT-ATG2 pathway controls mycobacterial infection. Nat. Commun. 8, 14642 doi: 10.1038/ncomms14642 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
O’Garra, A. et al. The immune response in tuberculosis. Annu. Rev. Immunol. 31, 475–527 (2013).
Cooper, A. M., Mayer-Barber, K. D. & Sher, A. Role of innate cytokines in mycobacterial infection. Mucosal Immunol. 4, 252–260 (2011).
Harris, J. et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity 27, 505–517 (2007).
O’Shea, J. J. & Plenge, R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 36, 542–550 (2012).
Redford, P. S., Murray, P. J. & O’Garra, A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 4, 261–270 (2011).
Wallis, R. S. & Hafner, R. Advancing host-directed therapy for tuberculosis. Nat. Rev. Immunol. 15, 255–263 (2015).
MacMicking, J. D. Cell-autonomous effector mechanisms against Mycobacterium tuberculosis. Cold Spring Harb. Perspect. Med. 4, a018507.
Lemaitre, B. & Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007).
Evans, C. J., Hartenstein, V. & Banerjee, U. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell 5, 673–690 (2003).
Clark, R. I., Woodcock, K. J., Geissmann, F., Trouillet, C. & Dionne, M. S. Multiple TGF-β superfamily signals modulate the adult Drosophila immune response. Curr. Biol. 21, 1672–1677 (2011).
Ganesan, S., Aggarwal, K., Paquette, N. & Silverman, N. NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr. Top. Microbiol. Immunol. 349, 25–60 (2011).
Dionne, M. S., Ghori, N. & Schneider, D. S. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71, 3540–3550 (2003).
Tobin, D. M. & Ramakrishnan, L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell. Microbiol. 10, 1027–1039 (2008).
Agaisse, H., Petersen, U. M., Boutros, M., Mathey-Prevot, B. & Perrimon, N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5, 441–450 (2003).
Oldefest, M. et al. Upd3--an ancestor of the four-helix bundle cytokines. Biochem. Biophys. Res. Commun. 436, 66–72 (2013).
Brown, S., Hu, N. & Hombria, J. C. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr. Biol. 11, 1700–1705 (2001).
Dostert, C. et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6, 946–953 (2005).
Krzemien, J. et al. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature 446, 325–328 (2007).
Buchon, N., Broderick, N. A., Chakrabarti, S. & Lemaitre, B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333–2344 (2009).
Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S. & Lemaitre, B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200–211 (2009).
Jiang, H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009).
Osman, D. et al. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell Sci. 125, 5944–5949 (2012).
Zhou, F., Rasmussen, A., Lee, S. & Agaisse, H. The UPD3 cytokine couples environmental challenge and intestinal stem cell division through modulation of JAK/STAT signaling in the stem cell microenvironment. Dev. Biol. 373, 383–393 (2013).
Wang, L. et al. Pleiotropy of the Drosophila JAK pathway cytokine Unpaired 3 in development and aging. Dev. Biol. 395, 218–231 (2014).
Olofsson, B. & Page, D. T. Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev. Biol. 279, 233–243 (2005).
Agaisse, H. & Perrimon, N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 198, 72–82 (2004).
Makki, R. et al. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 8, e1000441 (2010).
McGuire, S. E., Mao, Z. & Davis, R. L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 2004, pl6 (2004).
Simeone, R. et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 8, e1002507 (2012).
Behar, S. M. et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 4, 279–287 (2011).
Chen, M., Gan, H. & Remold, H. G. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J. Immunol. 176, 3707–3716 (2006).
Bina, S., Wright, V. M., Fisher, K. H., Milo, M. & Zeidler, M. P. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep. 11, 201–207 (2010).
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Singh, V. et al. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe 12, 669–681 (2012).
Velikkakath, A. K., Nishimura, T., Oita, E., Ishihara, N. & Mizushima, N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 23, 896–909 (2012).
Manzanillo, P. S. et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501, 512–516 (2013).
Peyron, P. et al. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 4, e1000204 (2008).
Russell, D. G., Cardona, P. J., Kim, M. J., Allain, S. & Altare, F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10, 943–948 (2009).
Daniel, J., Maamar, H., Deb, C., Sirakova, T. D. & Kolattukudy, P. E. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 7, e1002093 (2011).
Buszczak, M., Lu, X., Segraves, W. A., Chang, T. Y. & Cooley, L. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics 160, 1511–1518 (2002).
Nagabhushanam, V. et al. Innate inhibition of adaptive immunity: mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J. Immunol. 171, 4750–4757 (2003).
Martinez, A. N., Mehra, S. & Kaushal, D. Role of interleukin 6 in innate immunity to Mycobacterium tuberculosis infection. J. Infect. Dis. 207, 1253–1261 (2013).
Garton, N. J., Christensen, H., Minnikin, D. E., Adegbola, R. A. & Barer, M. R. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology 148, 2951–2958 (2002).
Bailey, A. P. et al. Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell 163, 340–353 (2015).
Anand, P. et al. A novel role for lipid droplets in the organismal antibacterial response. eLife 1, e00003 (2012).
Tobin, D. M. & Ramakrishnan, L. TB: the Yin and Yang of lipid mediators. Curr. Opin. Pharmacol. 13, 641–645 (2013).
Tobin, D. M. et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140, 717–730 (2010).
Kimmey, J. M. et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569 (2015).
Collins, C. A. et al. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 5, e1000430 (2009).
Flynn, J. L. & Chan, J. Tuberculosis: latency and reactivation. Infect. Immun. 69, 4195–4201 (2001).
Colas, C. et al. An improved flow cytometry assay to monitor phagosome acidification. J. Immunol. Methods 412, 1–13 (2014).
Kampmann, B. et al. Evaluation of human antimycobacterial immunity using recombinant reporter mycobacteria. J. Infect. Dis. 182, 895–901 (2000).
Renna, M. et al. Azithromycin blocks autophagy and may predispose cystic fibrosis patients to mycobacterial infection. J. Clin. Invest. 121, 3554–3563 (2011).
Sampson, S. L. et al. Protection elicited by a double leucine and pantothenate auxotroph of Mycobacterium tuberculosis in guinea pigs. Infect. Immun. 72, 3031–3037 (2004).
Schiebler, M. et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol. Med. 7, 127–139 (2015).
Floto, R. A. et al. Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science 314, 454–458 (2006).
Barth, J. M., Szabad, J., Hafen, E. & Kohler, K. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 18, 915–924 (2011).
Nagy, P. et al. Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy 10, 453–467 (2014).
Chan, K. et al. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc. Natl Acad. Sci. USA 99, 3920–3925 (2002).
Alland, D. et al. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 95, 13227–13232 (1998).
Dionne, M. S., Pham, L. N., Shirasu-Hiza, M. & Schneider, D. S. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr. Biol. 16, 1977–1985 (2006).
Acknowledgements
We thank Hervé gAaisse, James Castelli-Gair Hombría, Martin Zeidler, Bruno Lemaitre, Bruce Edgar, Teresa Żołądek, Lalita Ramakrishnan, the Vienna Drosophila RNAi Center and the Bloomington Drosophila Stock Center for flies or plasmids; Gabor Juhasz, Katja Köhler and Sara Cherry for antibodies to Drosophila ATG8; Brian Robertson, Bill Jacobs and Lalita Ramakrishnan for bacterial strains; Karen Liu, Serge Mostowy, Frédéric Geissmann, Chloë Colas, Katie Woodcock and members of the Dionne and Stramer laboratories for helpful discussion and comments. C.B.P. was supported by a PhD studentship from the Oliver Bird Rheumatism Programme. Early stages of this work were supported by grants from the BBSRC (BB/E02128X/1) and the Wellcome Trust (WT085119MA). Current work in the Dionne lab is funded by the BBSRC (BB/L020122/1) and MRC (MR/L018802/1).
Author information
Authors and Affiliations
Contributions
Study conception and design: M.S.D. and C.B.P. Acquisition of data: C.B.P., M.S., S.W.S.T., J.A.S., K.K., K.P.B., M.C.M., S.M., M.P., K.P.B. and M.S.D. Analysis and interpretation of data: C.B.P., M.S., P.G., B.M.S., R.A.F. and M.S.D. Drafting of manuscript: C.B.P. and M.S.D. Critical revision: M.S.D., R.A.F. and C.B.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures and Supplementary Table (PDF 3361 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Péan, C., Schiebler, M., Tan, S. et al. Regulation of phagocyte triglyceride by a STAT-ATG2 pathway controls mycobacterial infection. Nat Commun 8, 14642 (2017). https://doi.org/10.1038/ncomms14642
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms14642
This article is cited by
-
Mycobacterium abscessus pathogenesis identified by phenogenomic analyses
Nature Microbiology (2022)
-
Metabolic strategy of macrophages under homeostasis or immune stress in Drosophila
Marine Life Science & Technology (2022)
-
Emerging technologies and infection models in cellular microbiology
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.