Abstract
TH17 cells are recognized as a unique subset of T helper cells that have critical roles in the pathogenesis of autoimmunity and tissue inflammation. Although RORγt is necessary for the generation of TH17 cells, the molecular mechanisms underlying the functional diversity of TH17 cells are not fully understood. Here we show that a member of interferon regulatory factor (IRF) family of transcription factors, IRF8, has a critical role in silencing TH17-cell differentiation. Mice with a conventional knockout, as well as a T cell-specific deletion, of the Irf8 gene exhibited more efficient TH17 cells. Indeed, studies of an experimental model of colitis showed that IRF8 deficiency resulted in more severe inflammation with an enhanced TH17 phenotype. IRF8 was induced steadily and inhibited TH17-cell differentiation during TH17 lineage commitment at least in part through its physical interaction with RORγt. These findings define IRF8 as a novel intrinsic transcriptional inhibitor of TH17-cell differentiation.
Similar content being viewed by others
Introduction
CD4+ T helper (TH) T cell subsets are characterized by the secretion of unique cytokine profiles and have critical roles in orchestrating adaptive immune responses. In addition to TH1 and TH2 cells, TH17 cells have been identified more recently as a third TH subset mediating inflammatory and autoimmune responses through the production of interleukin (IL)-17A, IL-17F and IL-22 (refs 1, 2, 3, 4). TH17 lineage commitment is initially driven by transforming growth factor (TGF)-β in the presence of IL-6 or IL-21 (refs 5, 6, 7, 8), whereas IL-23 serves to expand or maintain TH17 populations2,5,9,10. The orphan nuclear receptor, RORC, also known as RORγt, has been identified as the master transcription factor for TH17 development11. The differentiation of TH17 cells is also regulated by several recently described positive and negative feedback loops involving IL-21, IL-23R, IL-10 and IL-27 (refs 6, 7, 12, 13, 14, 15), indicating that intrinsic genetic programmes may contribute to the silencing of TH17 lineage commitment. There is, increasing evidence that TH17 cells are involved in the pathogenesis of various autoimmune/inflammatory diseases, including multiple sclerosis, rheumatoid arthritis, inflammatory bowel diseases and asthma16. Thus, a more complete understating of the molecular mechanisms involved in the regulation of TH17 immune responses should provide insights into the pathogenesis and treatment of these and possibly other inflammatory diseases. Several transcription factors, including RORγt, RORα, STAT3 and interferon regulatory factor (IRF)4, have been reported to be important for TH17-cell differentiation. However, the silencing programme for TH17-cell differentiation has not been fully examined.
IRF8, a member of the IRF family, is expressed by B cells, dendritic cells (DCs), macrophages17,18,19 and activated T cells20,21, and has been shown to have a diverse roles in the regulation of innate and adaptive immune responses. IRF8 has a DNA-binding domain in the amino (N)-terminal half of the protein and an IRF association domain in the carboxy (C) terminus that is responsible for heterodimerization with other transcription factors22. IRF8 functions as a transcriptional repressor or activator depending on the formation of different heterodimeric DNA-binding complexes with partners that include members of the ETS family and the IRF family22. It is known that IRF8 has critical roles in the differentiation of myeloid cells, promoting monocyte over granulocyte differentiation23. It is also a crucial regulator of many aspects of DC development, differentiation and function24, thereby having an essential role in the establishment of innate immune responses. Although IRF8 is critical for the regulation of immune cell growth, differentiation and survival25, the direct effects of IRF8 on T-cell activation and differentiation are incompletely understood.
In the present study, we show that mice deficient in IRF8 because of a conventional knockout (KO) or with a T cell-specific conditional deletion exhibited enhanced TH17-cell differentiation while exhibiting no significant effects on TH1 or TH2 cells. In addition, transfer of naive T cells from IRF8-deficient mice induced more severe colitis in Rag–/– mice than T cell from normal controls. Furthermore, we report that IRF8 physically interacts with RORγt, resulting in inhibition of IL-17 transcription. These findings suggest that IRF8 has a suppressive role in the control of TH17 differentiation and highlight the importance of intrinsic genetic programmes for the silencing of TH17-dependent immune responses.
Results
IRF8 deficiency enhances TH17-cell differentiation
To investigate the function of IRF8 in T cells, we first examined the expression of IRF8 in CD4+ T cells from normal or OT-II transgenic mice activated by different stimuli. We found that T-cell antigen receptor (TCR) engagement with anti-CD3 and anti-CD28 antibodies as well as stimulation of OT-II cells resulted in significant induction of IRF8 protein expression, as determined by western blotting (Supplementary Fig. S1a,b). Interestingly, IRF8 protein was more stably expressed in naive CD4+ T cells polarized for 12 to 72 h under TH17-inducing conditions compared with TH1- or TH2-inducing conditions (Supplementary Fig. S1a). To clarify how TH17-polarizing conditions induce stable IRF8 expression, CD4+ cells were stimulated with TGF-β in the absence of TCR activation and the results showed that TGF-β clearly induced IRF8 expression at both 48 and 72 h (Supplementary Fig. S1c). In addition, mitogen-activated protein kinase inhibitors significantly blocked IRF8 protein expression induced by TCR activation (Supplementary Fig. S1d) and STAT3 mutant mice showed impaired IRF8 mRNA expression (Supplementary Fig. S1e). These data indicate that IRF8 is consistently elevated in activated CD4+ T cells and TCR signalling cascade is involved in induction of IRF8 expression.
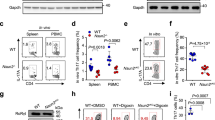
We then assessed the contributions of IRF8 to TH17 differentiation by studying CD4+ T cells from mice deficient in IRF8 due to a conventional KO of the gene (Irf8–/– mice). Naive CD4+ T cells from Irf8–/– or wild-type (WT) littermate mice were primed in vitro for 4 days under TH0 or TH17 polarizing conditions. The cells were then re-stimulated with phorbol myristate acetate (PMA)/ionomycin and examined for the percentages of IL-17-producing cells by intracellular staining using flow cytometry. Notably, the frequency of IL-17-producing cells generated from Irf8–/– T-cell cultures was about threefold greater than cells from WT cultures (Fig. 1a). These observations correlated with enhanced IL-17 secretion by Irf8–/– TH cells generated under TH17 polarizing conditions as determined by enzyme-linked immunosorbent assay (ELISA; Fig. 1b). In addition, IL-17-producing CD4+ T cells were significantly increased among lamina propria lymphocytes isolated from Irf8–/– mice as compared with WT littermate controls following in vitro activation under TH17 conditions or at the basal levels (Fig. 1c and Supplementary Fig. S2).
(a) Naive CD4+ T cells from wild-type (WT) or Irf8–/– mice were differentiated under TH0 and TH17 polarizing conditions for 4 days. Cells were then re-stimulated with PMA/ionomycin for 5 h, stained for intracellular IL-17 and IFN-γ, and analysed by flow cytometry. Representative fluorescence-activated cell sorting (FACS) dot plots gated on CD4+ cells and the percentages of IL-17-producing CD4+ cells are shown. Data are from one experiment representative of three independent experiments. Error bars, s.d. (b) The cells prepared in a, TH1 and TH2 polarizing conditions were re-stimulated with PMA/ionomycin for 24 h and the supernatants were analysed for IL-17 by ELISA. Data are from one experiment representative of three independent experiments. Error bars, s.d. (c) Wild-type or Irf8–/– lamina propria lymphocytes (LPL) were differentiated under TH17 conditions for 4 days. Cells were then re-stimulated with PMA/ionomycin for 5 h, stained for intracellular IL-17 and analysed by flow cytometry. (d) Naive CD4+ T cells from Lck-Cre+Irf8wt/wt and Lck-Cre+Irf8fl/fl mice were differentiated under TH0 and TH17 polarizing conditions for 4 days and the cells were re-stimulated with PMA/ionomycin for 5 h for staining of IL-17, IFN-γ and FOXP3. Representative FACS dot plots gated on CD4+ cells are shown. (e) The cells prepared in d were re-stimulated with PMA/ionomycin for 24 h and the supernatants were analysed for IL-17 by ELISA. Data are from one experiment representative of two independent experiments. Error bars, s.d. (f) Naive CD4+ T cells from C57BL/6 mice were differentiated under TH0 and TH17 conditions for 4 days. The cells were re-stimulated with PMA/ionomycin for 12 h and IRF8 expression was analysed by western blot. (g) The cells prepared in a were re-stimulated with PMA/ionomycin for 5 h and mRNA expression of indicated genes was determined by qPCR. The data shown were normalized to levels of ubiquitin expression as analysed by qPCR. The results are representative of three independent experiments. Error bars, s.d.
To determine whether the effects of IRF8 deficiency on TH17 potential were cell type-specific, we next compared CD4+ T cells from mice expressing Lck-Cre that were WT (wt/wt) or homozygous (fl/fl) for a conditional allele of Irf8 gene resulting in selective depletion of IRF8 in the T-cell compartment. Quantitative real-time reverse transcrption(RT)–PCR (qPCR) analyses revealed dramatically lower levels of Irf8 transcripts in sorted thymocyte sub-populations (DP, CD4SP, CD8SP) and splenic CD3+CD4+ cells from Lck-Cre+Irf8fl/fl compared with cells from Lck-Cre+Irf8wt/wt littermate control mice (Supplementary Fig. S3), confirming that the Irf8 gene was efficiently deleted from Lck-Cre+Irf8fl/fl T cells. Splenic and lymph node CD4+ T-cell subsets from mice with IRF8-deficient T cells as well as from mice homozygous for a conventional Irf8 null allele were normal in number as well as in expression of the T-cell activation markers CD62L, CD44, CD25 and CD69. Expression of FOXP3 in thymic and peripheral lymph node T cells from mice of both genotypes was also similar (Supplementary Fig. S4), indicating that CD4+ T cells develop normally in the absence of IRF8. Naive CD4+ T cells from Lck-Cre+Irf8fl/fl and Lck-Cre+Irf8wt/wt littermate controls were subjected to TH17 polarization. As expected, TH cells from mice with a T cell-specific deficiency in IRF8 showed a remarkable increase in the generation of IL-17-producing cells (Fig. 1d) in association with significantly elevated levels of IL-17 secretion (Fig. 1e). These results excluded the possibility that the effects of IRF8 deficiency on T cells from mice with a conventional KO of the gene could be attributed to the altered activities of other subsets of IRF8-deficient cells, such as B cells, DC or macrophages. TH17 cells generated from splenic T cells of Irf8–/– mice comprised a major portion of the β-TCR+ CD4+ lymphocyte subset (Supplementary Fig. S5). Indeed, high levels of IRF8 protein expression were detected in CD4+ T cells polarized under TH17 conditions as determined by immunoblot analyses (Fig. 1f). Accordingly, transcript levels of the iconic TH17 cytokines IL-17A, IL-17F and IL-22 were enhanced in Irf8–/– TH17 cells (Fig. 1g). In contrast, TH1 or TH2 differentiation was not noticeably affected in Irf8–/– T-cell cultures (Fig. 2a–c). In addition, [3H]-Thymidine incorporation assay showed that the proliferation of CD4+ T cells from Lck-Cre+Irf8fl/fl and Lck-Cre+Irf8wt/wt mice cultured under TH17 conditions was comparable (Fig. 2d). Taken together, these results indicate that IRF8 is induced during T-cell activation, and that TH17-cell differentiation is enhanced in cells deficient in IRF8.
Naive CD4+ T cells of WT or Irf8–/– mice were differentiated under TH1 or TH2 polarizing conditions for 4 days. Cells were then re-stimulated with PMA/ionomycin for 5 h and stained for intracellular IFN-γ as a TH1 marker (a) and IL-4 as a TH2 marker (b) by flow cytometry. Data are from one experiment representative of three independent experiments. Error bars, s.d. The cells prepared in a and b were re-stimulated with PMA/ionomycin for 12 h and the supernatants were analysed for IFN-γ and IL-4 by ELISA (c). Data are from one experiment representative of three independent experiments. Error bars, s.d. Naive CD4+ T cells from spleens and lymph nodes of Lck-Cre+Irf8wt/wt and Lck-Cre+Irf8fl/fl mice were prepared and the cells were differentiated under TH0 and TH17 polarizing conditions for 3 days. [3H]-Thymidine was added during the last 8 h of culture. Then the cells were collected and were counted with a beta-counter (d). Data are from one experiment representative of two independent experiments. Error bars, s.d.
Treg cells and autocrine cytokines are not altered in Irf8–/– mice
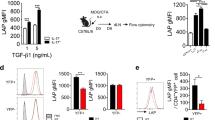
To understand whether alterations in Treg cells might contribute to enhanced TH17 differentiation in IRF8-deficient mice, we analysed FOXP3+ CD4+ T cells in these mice. There were no significant differences between the FOXP3+ CD4+ T-cell populations of WT and Irf8–/– mice under TH17- or Treg-inducing conditions (Fig. 3a,b). Thus, the more efficient generation of Irf8–/– TH17 cells in response to the combined effects of TGF-β plus IL-6 was not because of alterations in TGF-β-derived Treg suppression. IL-17-producing cells generated from Irf8–/– T-cell cultures were greatly increased following stimulation with IL-6 plus TGF-β (Fig. 3c).
Naive CD4+ T cells from wild-type and Irf8–/– mice were stimulated with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of IL-6 (10 ng ml−1) plus differing concentrations of TGF-β (0.1, 1, 5 ng ml−1) (a), or TGF-β (5 ng ml−1), TGF-β (5 ng ml−1)/IL-6 (10 ng ml−1) with or without retinoic acid (RA, 100 nM) (b). After 4 days of stimulation, IL-17 and FOXP3 intracellular staining was performed and analysed by flow cytometry. (c) The cells prepared in a and b except for the presence of TGF-β (5 ng ml−1) plus various concentrations of IL-6 (5, 10, 50, 100 ng ml−1) were re-stimulated with PMA/ionomycin for 5 h and stained for intracellular IL-17 and IFN-γ and analysed by flow cytometry. (d) Naive CD4+ T cells from wild-type and Irf8–/– mice were stimulated with IL-6, IL-23, TGF-β or different combinations of these cytokines for various intervals. Total RNA was extracted and analysed by RT–PCR for mRNA expression of IL-17 (top panel) and IL-17F (bottom panel). Data are from one experiment representative of three independent experiments. Error bars, s.d. (e) Naive CD4+ T cells from WT and Irf8–/– mice were stimulated with the indicated cytokines for 4 days. Cells were re-stimulated with PMA/ionomycin for 5 h and stained for intracellular IL-17 and IFN-γ and analysed by flow cytometry. The results are representative of three independent experiments. (f) Cells were prepared as in e and the culture supernatants were collected after 4 days of stimulation. IL-17 protein secretion was analysed by ELISA. (g) Naive CD4+ T cells from WT and Irf8–/– mice were prepared and stimulated with TGF-β and IL-6 in the presence of neutralizing anti-IFN-γ (10 μg ml−1) and anti-IL-4 (10 μg ml−1) antibodies. At 72 h after the stimulation, IL-17 protein secretion in culture supernatants was analysed by ELISA. Data are the mean±s.d. of triplicate cultures. (h) Naive CD4+ T cells from wild-type or Irf8–/– mice were stimulated with the indicated cytokine combinations for 48 h. Total RNA was extracted and analysed by qPCR for mRNA expression of the indicated genes. Data are from one experiment representative of three independent experiments. Error bars, s.d.
To further investigate how IRF8 affects TH17 differentiation, naive WT and Irf8–/– CD4+ T cells were subjected to TH17 differentiation in the presence of IL-6, IL-23 and TGF-β, either alone or in various combinations, and then examined for the expression of lineage-specific genes by qPCR. Neither IL-6 nor TGF-β alone induced significant levels of IL-17 or IL-17F transcripts (Fig. 3d). In contrast, dramatic increases in IL-17 and IL-17F transcripts were induced at multiple time points by the combination of IL-23, IL-6 and TGF-β in cultures of both Irf8–/– and WT T cells (Fig. 3d). Increased expression of IL-17 was confirmed at the protein level by ELISA (Fig. 3e–g). IL-23 induced low levels of IL-17 and IL-17F transcripts with no significant differences being seen between CD4+ T cells of WT and Irf8–/– mice (Fig. 3d). This suggests that IRF8 may not target the IL-23 signalling cascade, but may exert a major influence on TH17 differentiation instead of TH17 expansion and maintenance.
IL-21, an autocrine cytokine produced by CD4+ T follicular helper cells and TH17 cells6,7, induces TH17 differentiation in the presence of TGF-β. IRF8-deficient T cells displayed enhanced induction of TH17-associated molecules following stimulation by TGF-β combined with IL-6 or IL-21 (Figs 3h and 4a). However, production of IL-21 and IL-10 was comparable in WT and Irf8–/– TH17 cells (Fig. 4b–e). These results suggest that an autocrine loop involving either IL-21 or IL-10 is not involved in the functional control of TH17 differentiation by IRF8.
Naive CD4+ T cells from WT and Irf8–/– mice were prepared, and the cells were stimulated with anti-CD3 and anti-CD28 antibodies in the presence or absence of TGF-β plus IL-21 or IL-6 for 72 h. Intracellular staining for IL-17 and IFN-γ expression was performed and analysed by flow cytometry (a). The secretion of IL-21 protein in culture supernatants was analysed by ELISA (b). Data are from one experiment representative of three independent experiments. Error bars, s.d. Naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 antibodies in the presence of TGF-β plus IL-6 or IL-21. At 96 h after stimulation, intracellular staining for IL-17 and IL-10 was performed and analysed after re-stimulation with PMA/ionomycin by flow cytometry (c). Naive CD4+ T cells from spleens and lymph nodes of WT and Irf8–/– mice were prepared and the cells were stimulated under TH0, TH1, TH2 and TH17 polarizing conditions for 4 days. Then the cells were re-stimulated with PMA/ionomycin for 12 h and IL-10 protein secretion was analysed by ELISA (d). Data are from one experiment representative of three independent experiments. Error bars, s.d. Naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 antibodies in the presence of TGF-β plus IL-6 or IL-21 for 72 h. IL-10 expression was analysed by RT–PCR (e). Data are from one experiment representative of three independent experiments. Error bars, s.d.
We next determined whether the induction of TH17-associated genes may be affected by forced expression of IRF8 in T cells. Retroviral transduction of IRF8-IRES-GFP into WT naive CD4+ T cells significantly decreased the percentage of IL-17-producing cells under TH17 polarizing conditions (Fig. 5a), and RORγt-positive cells were moderately reduced (Fig. 5a). Similarly, retroviral transduction of IRF8 into the EL4T lymphoma cell line stimulated with PMA/ionomycin resulted in significantly reduced transcripts for IL-17, but had no effect on the expression of interferon (IFN)-γ, IL-4, IL-10 or FOXP3 (Fig. 5b). Taken together, these results demonstrate a direct role for IRF8 in suppressing TH17-specific gene expression.
(a) Naive CD4+ T cells from C57BL/6 mice were infected with retrovirus encoding IRF8 or empty vector and were activated under TH17-inducing conditions for 4 days. The cells were re-stimulated with PMA/ionomycin for 5 h and stained for intracellular IL-17 and RORγt and analysed by flow cytometry. (b) EL4 cells were transduced with retroviruses encoding IRF8 (black column) or GFP (white column) for 48 h and the transduced cells were then stimulated with PMA/ionomycin for various times as indicated. Total RNA was extracted and the transcript levels of TH17-associated genes were analysed by qPCR as indicated. The results are normalized to ubiquitin levels. Data are from one experiment representative of three independent experiments. Error bars, s.d.
IRF8 interacts with RORγt and suppresses IL-17 transcription
The above findings prompted us to probe the molecular basis for IRF8 control of TH17-cell differentiation. As many studies have demonstrated a critical role for RORγt in TH17-cell differentiation both in vitro and in vivo11, we asked if IRF8 might affect RORγt-mediated IL-17 induction. EL4 cells were transiently transfected RORγt followed by stimulation with PMA/ionomycin. Overexpression of RORγt resulted in significantly increased expression of IL-17 and IL-17F, whereas co-transfection with IRF8 greatly reduced the expression of these genes, suggesting that IRF8 inhibits RORγt-induced expression of IL-17 transcripts (Fig. 6a). Using a 6-kbp IL-17 promoter reporter plasmid, we confirmed that RORγt strongly induced IL-17 promoter reporter activity in 293T cells (Fig. 6b), which was shown to be due to a direct effect of RORγt on the IL-17 promoter26,27. Co-transfection of IRF8 in these cells suppressed RORγt-mediated IL-17 promoter activity in a dose-dependent manner (Fig. 6c). Similar results were observed in EL4 cells (Fig. 6d). CNS2 is a conserved non-coding sequence (CNS) element ∼5 kbp upstream of the IL-17 locus that functions as a RORγt-dependent enhancer element required for optimal IL-17 transcription27,28. Overexpression of IRF8 significantly suppressed CNS2-enhanced IL-17 promoter activity (Fig. 6e), indicating that IRF8 inhibits IL-17 transcription.
(a) IRF8-expressing EL4 cells (black column) or control cells (white column) were transiently transfected with a RORγt plasmid for 24 h, and the cells were treated with plate-bound anti-CD3 and anti-CD28 antibodies in the presence or absence of the indicated cytokines for 8 h. qPCR analyses of transcripts of the indicated genes were performed and the results were normalized to the levels of ubiquitin transcripts. **P<0.01 versus cells transfected with IRF8 (Student's t-test). Data are representative of multiple experiments. (b) 293T cells were co-transfected with an IL-17 promoter reporter construct containing a 6-kbp promoter and increasing doses of a RORγt plasmid for 30 h. (c) 293T cells were co-transfected with an IL-17 promoter reporter construct containing a 6-kbp promoter, a RORγt plasmid, a FOXP3 plasmid, and increasing doses of an IRF8 plasmid for 30 h. (d) IRF8-expressing EL4 cells (black column) or control cells (white column) were co-transfected with an IL-17 promoter reporter construct containing a 6-kbp promoter plus RORγt plasmid, and treated with plate-bound anti-CD3 antibody in the presence or absence of the indicated cytokines for 24 h. Luciferase activities in (b, c, d) were measured and normalized to β-galactosidase activity. Data are mean±s.d. of triplicate cultures and are representative of three independent experiments. *P<0.05 versus cells transfected with IRF8 (Student's t-test). (e) IRF8-expressing EL4 cells (black column) or control cells (white column) were co-transfected with an IL-17 promoter reporter construct containing a minimal 1.1-kb promoter plus CNS2, and RORγt plasmids. The cells were treated with plate-bound anti-CD3 and anti-CD28 antibodies and IL-6/TGF-β for 24 h. A luciferase assay was performed as described in (b), (c), and (d). *P<0.05 versus cells transfected with IRF8 (Student's t-test). (f) 293T cells were co-transfected with an IL-17 promoter reporter construct (containing CNS2) with either an IRF8 overexpression plasmid or a control vector for 36 h, followed by ChIP analysis. 3 μg of anti-IRF8 antibody or isotype-matched IgG as control antibody were used in the immunoprecipitation step. qPCR was used to quantify the amount of precipitated DNA with primers flanking the CNS2 region of the IL-17 promoter. Data were normalized to input DNA in each respective sample. (g) Naive CD4+ T cells from wild-type or Irf8–/– mice were cultured under TH17-polarizing conditions for 60 h, followed by ChIP assay as described above. 3 μg of anti-IRF8 antibody or isotype-matched IgG as control antibody were used in the immunoprecipitation step. PCR was used to quantify the amount of precipitated DNA with primers flanking the CNS2 region of the IL-17 promoter.
To investigate whether IRF8 can directly bind the CNS2 region of the IL-17 promoter, we co-transfected an IL-17 promoter reporter (containing CNS2) and IRF8 plasmids into 293T cells and performed chromatin immunoprecipitation (ChIP) using an IRF8-specific antibody. The precipitated chromatin DNA was analysed by qPCR using primers covering the CNS2 region of the IL-17 promoter. The results showed that IRF8 antibody specifically pulled down the CNS2 region sequences (Fig. 6f). To confirm the results, naive WT and Irf8–/– CD4+ T cells were stimulated under TH17 polarizing conditions for 60 h and ChIP assay was performed as above. The precipitated chromatin DNA was analysed by PCR using primers covering the CNS2 region of the IL-17 promoter. Similarly, IRF8 antibody specifically pulled down the CNS2 region sequences of TH17 cells from WT mice but not from Irf8–/– mice (Fig. 6g). These results demonstrate that IRF8 bound directly to this region of the IL-17 promoter. Further analyses of the CNS2 sequence revealed several IRF consensus binding sequence elements (Supplementary Fig. S6a). We showed that mutation of a typical IRF-binding site in the CNS2 region of IL-17 promoter nullified the inhibition of RORγt-mediated IL-17 promoter activation by IRF8 (Supplementary Fig. S6b). These results provided a molecular basis for understanding the inhibitory effects of IRF8 on IL-17 transcription. It is likely that RORγt cooperates with other known transcription factors, such as FOXP3 or RUNX1, or unknown factors in the coordinate regulation of TH17 differentiation26,29,30. We then co-transfected HA-tagged IRF8 and T7-RORγt plasmids into 293T cells for co-immunoprecipitation. Immunoprecipitation of RORγt resulted in co-precipitation of IRF8 (Fig. 7a), even in the presence of ethidium bromide or DNase I, indicating that IRF8 and RORγt interact with each other without the involvement of DNA. Using confocal microscopy, we also determined that IRF8 and RORγt co-localized in the nucleus of NIH3T3 cells co-transfected with GFP-IRF8 and T7-RORγt constructs (Fig. 7b). Flow cytometric analyses of naive CD4+ T cells stimulated under TH17 polarizing conditions clearly revealed a population of RORγt+CD4+ cells with the majority of these cells also staining for IRF8 (Fig. 7c). Furthermore, anti-IRF8 antibodies were found to co-immunoprecipitate RORγt from lysates of primary CD4+ T cells from WT but not from Irf8–/– mice cultured under TH17 polarizing conditions (Fig. 7d), indicating that endogenous IRF8 and RORγt interact with each other. As shown in Figure 7e, IRF8 is comprised of an N-terminal DNA-binding domain, a flanking internal region, and a C-terminal IRF association domain31. To map the binding sites between IRF8 on RORγt, we co-transfected 293T cells with T7-tagged RORγt and Flag-tagged full-length IRF8 or one of a series of C-terminal truncation mutants (1–390, 1–356, 1–305, 1–253, 1–230, 1–190 and 1–154) followed by co-immunoprecipitation experiments. The results showed that IRF8 amino-acid residues between 230 and 190 were important for the physical interaction with RORγt (Supplementary Fig. S7a, Fig. 7f). IRF8 mutant (1–114), which is not bound to RORγt, did not suppress RORγt-mediated IL-17 promoter activation as WT and other binding mutants did (Fig. 7g and Supplementary Fig. S7b), demonstrating that the inhibitory activity of IRF8 on IL-17 transcription is related to its interaction with RORγt. In addition, IRF4, another IRF family member that is required for the generation of TH17 cells32, was also found in co-immunoprecipitation experiments to interact with RORγt, whereas there was no interaction between IRF1 and RORγt (Supplementary Fig. S8a). Although transcript levels for IRF4 in cells deficient in IRF8 did not change under TH17 polarizing conditions (Supplementary Fig. S8b), we still cannot exclude possibility that IRF8 and IRF4 mutually influence their activities in TH17 cells. Thus, our results suggest that protein–protein interactions between IRF8 and RORγt have at least a partial role in IRF8-mediated inhibitory effects on IL-17 transcription.
(a) 293T cells were transfected with HA-tagged IRF8 and T7-tagged RORγt overexpression plasmids for 40 h and cell lysates were prepared in the presence or absence of DNase I or ethidium bromide. 500 μg of cell lysates were immunoprecipitated with an anti-T7 antibody and immunoblotted with specific antibodies as indicated. Data are representative of three independent experiments. (b) NIH3T3 cells were transiently transfected with GFP-IRF8 and T7-RORγt for 40 h, and cells were fixed and stained red for RORγt followed by confocal microscopic analysis. Scale bar, 50 μm. (c) Naive CD4+ T cells from WT mice were cultured under TH17-polarizing conditions for 72 h and the expression of RORγt and IRF8 was analysed by flow cytometry. The cells were gated on CD4+ T cells. Data are representative of three independent experiments. (d) Naive CD4+ T cells from wild-type or Irf8–/– mice were cultured under TH17-polarizing conditions for 60 h and the cell lysates were then immunoprecipitated with an anti-IRF8 antibody and western blotted (WB) with anti-RORγt and anti-IRF8 antibodies. Data represent three independent experiments. (e) Diagrams of IRF8 protein domains. (f) 293T cells were co-transfected with plasmids containing Flag-tagged full-length IRF8, IRF8 fragments (1–230, 1–190, 1–154) and T7-tagged RORγt plasmid for 40 h, and co-immunoprecipitation using anti-Flag antibody from the cell extracts was performed and immunoblotted with anti-T7 antibody. Data are representative of three independent experiments. (g) 293T cells were co-transfected with an IL-17 promoter reporter construct containing the 6-kbp promoter, a RORγt plasmid and either a full-length IRF8 or the IRF8 truncation mutant (1–114) construct for 30 h. Luciferase assays were performed as described in b. Data indicate mean±s.d. of triplicate cultures and are representative of three independent experiments. *P<0.05 versus cells transfected with IRF8 mutant (Student's t-test).
IRF8 controls TH17-cell differentiation in vivo
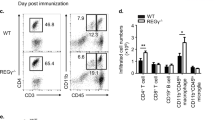
To further assess the effects of IRF8 on TH17-cell differentiation in vivo, we performed adoptive transfer experiments using CD4+CD62L+CD45RBhiCD25– cells from WT and Irf8–/– mice to induce colitis in RAG1 KO (Rag1–/–) mice. Irf8–/– mice did not develop spontaneous colitis during an observation period of 1.5 years (Fig. 8a,b). However, Rag1–/– mice reconstituted with Irf8–/– naive CD4+ T cells began losing weight earlier and lost more weight than mice in the control group. Parallel histological studies of colon sections from Rag1–/– mice reconstituted with Irf8–/– T cells revealed more severe inflammatory cell infiltrates and significantly higher pathological scores than those observed in sections from mice reconstituted with T cells from WT mice (Fig. 8c–e). In addition, mice reconstituted with Irf8–/– cells had a significantly higher percentage of IL-17-producing cells than control mice (Fig. 8f). To determine whether Treg cells from Irf8–/– mice could suppress effector T cells, we examined the population of Treg cells in Rag1–/– mice after the transfer of naive WT or Irf8–/– CD4+ T cells. Rag1–/– recipients of naive CD4+ T cells from mice of either genotype generated a small percentage of CD4+FOXP3+ and CD4+IFN-γ+ cells in the mesenteric lymph nodes (Fig. 8f). Furthermore, co-transfers of naive WT CD4+ T cells with CD4+CD25+ cells purified from WT or Irf8–/– mice resulted in similar effects on body weight (Supplementary Fig. S9). These results indicate that the effects of IRF8 deficiency on T cell-mediated inflammation could not be explained by influences on the function of CD4+ Treg cells. Thus, IRF8 deficiency promotes intestinal inflammation in a T cell-mediated model of colitis, suggesting that IRF8 may have an inhibitory role in the control of TH17-mediated immune responses. To further understand the role of IRF8 in TH17-cell differentiation in vivo, we extended our observations to an infection model, as TH17 cells have also been proposed to have a role in inflammation against both intracellular and extracellular bacteria33,34. Staphylococcus aureus is a Gram-positive bacterium that can induce IL-17 production from CD4+ T cells mainly through Staphylococcus aureus enterotoxin A (SEA), and humans deficient in TH17 cells are highly susceptible to infection with this agent35,36. To better understand the regulatory effects of IRF8 on TH17-cell differentiation in a broader sense, we used superantigenic S. aureus to induce IL-17 production in vivo. Spleen cells from Lck-Cre+Irf8fl/fl and Lck-Cre+Irf8wt/wt littermate controls immunized with SEA 4 days previously were re-stimulated in vitro with SEA for an additional 2 days and then examined for IL-17-producing CD4+ T cells by flow cytometry and for IL-17 secretion by ELISA (Supplementary Fig. S10). IRF8-deficient mice generated significantly more IL-17-producing CD4+ T cells than WT mice, further confirming that IRF8 negatively regulates TH17-cell differentiation in vivo.
WT and Irf8–/– mice were maintained under specific pathogen free (SPF). conditions for up to 18 months. Mice were killed and intestines were removed for histological analysis. Histology of colon tissues (a) and disease score (b) from age-matched young (15 weeks) and old (17–18 months) WT and Irf8–/– mice (three to four mice in each group). Scale bars, 200 μm. CD4+CD45RBhi T cells were purified from spleens and lymph nodes of wild-type or Irf8–/– mice and 5×105 cells were injected (i.p.) into recipient Rag–/– mice. Body weight change was monitored every week and mice were killed 7 weeks later. (c) Changes in body weight of Rag1–/– mice (n=5–6 mice per group) after intraperitoneal transfer of WT or Irf8–/– CD4+CD45RBhi T cells were recorded. Data are presented as the mean±s.d. of the percentage of initial body weight and are representative of two similar experiments. *P<0.05 versus recipients of WT cells (ANOVA test and Student's t-test). Disease scores (d) and sections of colons with colitis (e) from Rag1–/– mice (n=5–6 mice in each group) on day 35 after naive T cell transfer as described in c. *P<0.05 versus recipients of WT cells (Mann–Whitney test). Scale bars, 200 μm. (f) The percentage of IL-17, IFN-γ and FOXP3-producing cells from mesenteric lymph nodes of Rag1–/– mice in c (white column, transfer with WT cells; black column, transfer with Irf8–/– cells). **P<0.01 versus wild-type cell transferred mice (Student's t-test). Data are presented as the mean±s.d. from four mice in each group. Two independent experiments were performed with similar results.
Discussion
TH17 cells represent a recently defined member of a still growing family of T helper cells. The mechanisms involved in the silencing programme for this T helper subset remain unclear. Here we demonstrate that IRF8 serves as an intrinsic silencer for TH17-cell differentiation. IRF8-deficiency in both conventional and T cell-specific conditional KO mice led to more robust TH17-cell differentiation without effects on either TH1 or TH2 cell lineages. Furthermore, transfer of IRF8–/– CD4+CD45Rbhi cells into Rag1–/– mice induced more severe colitis than transfer of WT CD4+CD45Rbhi cells. In addition, mice reconstituted with IRF8–/– cells had a significantly higher percentage of IL-17-producing cells than mice reconstituted with WT cells. In addition, we showed that IRF8 physically interacts with RORγt resulting in suppression of IL-17 transcription. These results suggest that IRF8 negatively regulates the development of TH17 immune response resulting in the control of inflammation.
Many studies have demonstrated that IRF8 has important functions in myeloid cells24. Macrophages from Irf8–/– mice did not produce IL-12 in response to IFN-γ and LPS. IRF8 regulates IL-12 expression by binding to the IL-12 p40 promoter region, acting in synergy with IRF1 to activate IL-12 p40 gene expression37. In addition, IRF8 also induces the expression of other inflammatory proteins expressed by myeloid cells, including iNOS, IL-18 and IL-1 (refs 38, 39, 40). IRF8 protein levels are controlled in part by Cbl-mediated ubiquitylation and subsequent proteasomal degradation41. More recent studies have shown that transcriptional activation of IL-12p40 by IRF8 is enhanced following ubiquitylation by the E3 ubiquitin ligase, TRIM21 (ref. 42). Here, we showed that CD4+ T cells clearly expressed IRF8 protein on TCR engagement and that TH17 polarization conditions induced stable IRF8 protein expression. In addition, mitogen-activated protein kinase inhibitors completely blocked IRF8 protein expression induced by TCR activation and STAT3 mutant mice showed impaired IRF8 mRNA expression, indicating that the TCR signalling cascade is involved in induction of IRF8 expression. The percentages of CD4+ T cells in tissues of Irf8–/– and WT mice were comparable. Following stimulation under TH17 polarizing conditions, however, the percentages of IL-17-producing CD4+ T cells were greatly increased and the expression of TH17 signature genes was significantly enhanced for cells from Irf8–/– as compared with WT mice. These results suggest that IRF8 is an important transcription factor in controlling CD4+ T cell plasticity by targeting TH17-cell differentiation.
The balance between pathogenic TH17 cells and suppressive Treg cells in the immune system depends on the presence of inflammatory cytokines such as IL-6 and IL-21 (ref. 43). The anti-inflammatory cytokine, TGF-β, combined with IL-6 or IL-21 can drive the conversion of a TH cell phenotype from TGF-β-induced FOXP3-expressing Treg cells to RORγt-expressing TH17 cells. TGF-β inhibits the expression of STAT4 and GATA3, thereby preventing the differentiation of TH1 and TH2 cells, respectively, and concurrently facilitating TH17-cell development. CD4+ T cells from WT and Irf8–/– mice yielded similar populations of FOXP3+ cells following stimulation under TH17- or Treg-inducing conditions. In addition, there were no significant differences between WT and Irf8–/– TH17 cells in the production of the autocrine cytokines, IL-21 and IL-10. These results rule out the possibility that enhanced generation of TH17 cells by CD4+ T cells from Irf8–/– mice stimulated with TGF-β plus IL-6 was due to alterations in TGF-β-derived Treg suppression or to an autocrine loop involving IL-21 or IL-10.
IRF8 acts as a transcriptional repressor or a transcriptional activator depending on the target DNA sequence and interactions with different partner proteins, including PU.1, E47 and other IRFs25. We demonstrated that IRF8 interacts directly with RORγt, resulting in suppression of IL-17 transcription. The association of IRF8 with RORγt was not competed by FOXP3, another RORγt-binding protein44, indicating that IRF8 antagonizes the effect of RORγt without the involvement of FOXP3. In addition, co-immunoprecipitation studies showed that IRF4, another IRF family member required for TH17-cell differentiation32, also interacts with RORγt. It is likely that RORγt cooperates with other transcription factors, such as FOXP3 or RUNX1, or unknown factors in the regulation of TH17 differentiation26,29,30. It remains to be determined how these factors might collaborate with RORγt, the master transcription factor for TH17 cells to regulate differentiation of this TH subset.
TH17 cells are critical pathogenic effector T cells in inflammatory disorders such as inflammatory bowel disease45,46,47. In the present study, we demonstrated that IRF8 targets RORγt, resulting in the silencing of TH17-cell differentiation. In addition, transfer of IRF8–/– CD4+CD45Rbhi cells into Rag1–/– mice induced more severe colitis than transfer of WT cells. These results suggest that IRF8 functions as an important transcription factor in the control of inflammation by modulating RORγt activity. A recent genome-wide association study identifying IRF8 as a susceptibility locus in patients with multiple sclerosis48 is supportive of this model for IRF8 function in inflammatory diseases. As a result, our data may provide a molecular basis for identifying specific single-nucleotide polymorphisms associated with susceptibility to clinical immune pathologies.
Taken together, our results demonstrate that IRF8 is stably expressed during TH17-cell differentiation and has a critical role in directing the silencing programme for TH17-cell development. On the basis of these studies, we propose a novel molecular mechanism for the inhibitory effects of IRF8 on TH17 differentiation and cytokine expression that involves the modulation of RORγt activity (Supplementary Fig. S11). Our observations support a pathogenic role for TH17 cells in exacerbating inflammation and indicate that IRF8 may be a therapeutic target for controlling TH17-mediated autoimmune and inflammatory diseases.
Methods
Mice
C57BL/6 (B6) and B6-Irf8–/– mice were maintained in the barrier facility at the Mount Sinai School of Medicine. GFP-FOXP3 mice were crossbred with Irf8–/– mice to obtain Irf8–/–GFP-FOXP3 mice. IRF8 conditional knockout mice (Irf8fl/fl) were generated at Ozgene under a contract with NIAID by flanking exon 2 and an inserted PGK-neo cassette with loxP sites. Following homologous recombination of the targeting vector in C57BL/6 ES cells and establishment of germ line transmission, the PKG-neo cassette, which was flanked by flippase recognition target (FRT) sites, was excised by crossing with a FLP transgenic mouse. Selective breeding was used to eliminate the FLP gene. Conditional deletion of IRF8 in T cells was performed by crossing with Lck-Cre mice to generate Lck-Cre+Irf8fl/fl mice. The animal study protocols were approved by the Institutional Animal Care and Use Committees of Mount Sinai, NICHD and NIAID (protocol LIP-4).
Antibodies
The following antibodies were purchased from BD Biosciences, as conjugated to FITC, PE, PE-Cy5, perCP-Cy5.5 or APC: CD4 (L3T4), CD8 (53-6.7), CD3e (145-2C11), CD25 (PC61.5), CD44 (IM7), CD62L (MEL-14), CD45RB (C363-16A), IL-17 (TC11-18H10), IFN-γ (XMG1.2), TCRβ chain (H57-597) and isotype controls. Antibodies for IL-2 (JES6-1A12), IL-4 (11B11), IL-10 (JES5-16E3) and Foxp3 (FJK-16S) were purchased from eBiosciences.
CD4+ T cell preparation and differentiation in vitro
Naive CD4+ T cells (CD62L+CD44lo) were prepared by fluorescence-activated cell sorting from spleens and lymph nodes of Irf8–/– and WT littermates. The sorted cells were primed for 96 h with anti-CD3 (1 μg ml−1; 145-2C11; BD Biosciences) and soluble anti-CD28 (2 μg ml−1; 37.51; BD Biosciences). The cells were rested for 48 h, and were then re-stimulated for 5 h with PMA plus ionomycin in the presence of brefeldin A and intracellular cytokines were measured by flow cytometry. Cells stimulated under neutral conditions were defined as TH0 cells. Cells were stimulated to differentiate into TH1 cells by supplementation with IL-12 plus anti-IL-4 (10 μg ml−1; 11B11; BD Bioscences) or into TH2 cells by supplementation with IL-4 and anti-IFN-γ (10 μg ml−1; XMG1.2, BD Biosciences). For TH17-cell differentiation, cells were stimulated with transforming growth factor-β1 (5 ng ml−1), IL-6 (20 ng ml−1) and IL-23 (10 ng ml−1; all from R&D Systems) in the presence of anti-IL-4 antibody (10 μg ml−1; 11B11, BD Bioscences) and anti-IFN-γ antibody (10 μg ml−1; XMG1.2, BD Biosciences).
Intracellular staining and flow cytometry
Cells were stimulated with PMA and ionomycin for 5 h in the presence of brefeldin A before intracellular staining. Cells were fixed with IC Fixation Buffer (BD Biosciences), incubated with permeabilization buffer, and stained with PE-anti-mouse IL-17, APC-anti-IFN-γ and PE-Cy 5.5 anti-mouse CD4 antibodies. Flow cytometry was performed on a FACSCalibur (BD Biosciences) and LSR Fortessa (BD Biosciences).
RNA isolation and quantitative real-time RT–PCR
Total RNA was extracted using an RNeasy plus kit (QIAGEN) and cDNA was generated with an oligo (dT) primer and the Superscript II system (Invitrogen) followed by analysis using iCycler PCR with SYBR Green PCR master Mix (Applied Biosystems). Results were normalized based on the expression of ubiquitin. The following primer sets were used: IL-17A, IL-17F, IL-21, IL-23R, RORγt, IFN-γ, IRF8, IRF1 IRF4 T-bet FOXP3, CCR6, IL-10, IL-4, IL-2 and ubiquitin (Supplementary Table S1).
Transfection and luciferase reporter assay
293T cells were transiently transfected with an IL-17 promoter luciferase reporter plasmid together with RORγt in the presence of IRF8 plasmid at different concentrations. For each transfection, 2.0 μg of plasmid was mixed with 100 μl of Opti-MEM I medium (without serum and antibiotics) and 4.0 μl of Lipofectamine 2000 reagent. The mixture was incubated at room temperature for 20 min and added to 12-well plates containing cells and complete medium. The cells were incubated for 30 h and collected using reporter lysis buffer (Promega) for determination of luciferase activity. Cells were co-transfected with a β-galactosidase reporter plasmid to normalize experiments for transfection efficiency.
Generation of the mutant IL-17 promoters
A predicted IRF-binding site adjacent to the downstream RORγt-binding site in the CNS2 region of the mouse IL-17A promoter was mutated using QuickChange XL Site-Directed Mutagenesis Kit (Stratagene) according to manufacture's instruction. Two mutations were introduced on the IL-17 promoter regions −5204 to −5202 (TGG to CCC) and −5201 to −5199 (AAA to CCC) from the transcriptional initiation site (+1) using the following primer sets: 5′-GGTTGGAAAAAAAAACCCAAAGTTTTCTGACCCA-3′ and 5′-TGGGTCAGAAAACTTGGGTTTTTTTTTCCAACC-3′ for TGG to CCC; 5′-TGGAAAAAAAAATGGCCCGTTTTCT-GACCCACT-3′ and 5′-AGTGGGTCAGAAAACGGGCCATTTTTTTTTCCA-3′ for AAA to CCC. Mutations were verified by sequencing.
Retroviral transduction of IRF8 in CD4+ T cells
To prepare pseudotyped virus human 293 EbnaT cells were seeded at a density of 4×106 cells in a 10-cm dish. The next day, cells were transfected using calcium phosphate with a mixture of 2.5 μg of plasmid pMD.G encoding vesicular stomatitis virus G protein, 7.5 μg of plasmid encoding gag-pol, and 10 μg of a retroviral expression constructs encoding GFP or IRF8. At 48 h after transfection, the viral supernatant was collected, centrifuged at 800g, and used to infect cells. CD4+ T-cell transduction was performed as previously described11, sorted naive CD4+ T cells were plated as above and cultured for 24 h in the presence of anti-CD3 (1 μg ml−1; 145-2C11, BD Biosciences) and soluble anti-CD28 (2 μg ml−1; 37.51; BD Biosciences). Activated cells were then transduced with fresh retrovirus supernatant by centrifugation for 1.5 h at 2000g in the presence of polybrene (6 μg ml−1; Sigma). After 24 h, the cells were re-transduced using the same procedure and cultured for an additional 24 h and were then stimulated with TGF-β and IL-6. The cells were collected on day 5 or 6 for intracellular cytokine staining.
T-cell proliferation assay
Naive CD4+ T cells were purified from spleens and lymph nodes of Lck-Cre+Irf8fl/fl and Lck-Cre+Irf8wt/wt littermate controls. Cells (1×105 per well) were cultured in the absence or presence of anti-CD3 (1 μg ml−1) and anti-CD28 (2 μg ml−1) antibodies for 3 days in 96-well microplates. [3H]-Thymidine was added during the last 8 h of a 72-h culture. The cells were then collected and counted with a beta-counter.
Co-immunoprecipitation and immunoblotting analysis
Cells were washed with cold phosphate-buffered saline and lysed for 15 min on ice in 0.5 ml of lysis buffer (50 mM Tris-HCl, pH 8.0, 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol and 1 mM dithiothreitol) containing protease inhibitors. Cell lysates were clarified by centrifugation (4 °C, 15 min, 20,000 r.p.m.), aliquots (500 μg) were incubated with 2 μg of normal rabbit IgG for 4 h and 20 μl of protein G-Sepharose was added to the mixture for 2 h. After centrifugation, the supernatant was collected and incubated with 2 μg of anti-Flag antibody overnight at 4 °C with gentle rocking, after which immune complexes were collected as described above with 20 μl of protein G-Sepharose. After washing five times with lysis buffer, immunoblotting was performed. Anti-Flag (Sigma), anti-IRF8 (Santa Cruz), anti-β-actin (Sigma) and anti-T7 (MBL) antibodies were used according to the manufactures' instructions. Secondary antibodies were from Santa Cruz.
T-cell-transfer colitis studies and histopathology
T-cell-transfer colitis was performed as previously described49,50. Briefly, purified CD4+CD45RBhi T cells from WT and Irf8–/– mice were injected intraperitoneally into Rag1–/– recipients (5×105 cells per mouse in 200 μl sterile PBS per injection). Mice were weighed every week throughout the course of experiments. After 5 weeks, mice were killed and colon tissues were excised. Tissues were fixed in 10% buffered formalin and paraffin embedded. The sections (5 μm) of tissue samples stained with hematoxylin and eosin. All the slides were read and scored by an experienced pathologist (L.Q.) without previous knowledge of the type of treatment. The degree of inflammation in the epithelium, submucosa and muscularis propria was scored separately as described by Totsuka et al.49
In vivo stimulation of TH17 cells by Staphylococcus aureus
Lck-Cre+Irf8fl/fl mice and Lck-Cre+Irf8wt/wtl littermate controls were immunized (i.p.) with SEA (10 μg per mouse) for 4 days. Mice were killed and spleen cells were prepared. The cells were re-stimulated in vitro with SEA (10 ng ml−1) for additional 2 days and cells were collected for the analysis of IL-17-producing CD4+ T cells by flow cytometry gating on CD4+ T cells. The supernatants were collected for the measurement of IL-17 production by ELISA.
Chromatin immunoprecipitation assay
The ChIP procedure was performed using an assay kit following the manufacturer's instruction (Upstate Biotechnology). Briefly, 293T cells were co-transfected with an IL-17 promoter containing CNS2 domain and an IRF8 plasmid for 36 h and 1×107 transfected cells were then crosslinked by 1% formaldehyde for 10 min at 37 °C. Nuclei were prepared and subjected to sonication to obtain DNA fragments. Chromatin fractions were precleared with protein A-agarose beads followed by immunoprecipitation overnight at 4 °C with 3 μg of anti-IRF8 or control antibody. Crosslinking was reversed at 65 °C for 4 h, followed by proteinase K digestion. DNA was purified and subjected to qPCR. The input DNA was diluted 200 times before PCR amplification. The input and immunoprecipitated DNA were amplified by qPCR using primers (5′-CAGCCCTGGTCCTTAAACTG-3′ and 5′-TCACTTTCGTTGTGCCTTTG-3′) encompassing the CNS2 region of the mouse IL-17 promoter.
Cytokine ELISA
Supernatants from cell cultures were collected after activation under various conditions and secreted cytokines in the supernatants were measured by ELISA kits with purified coating and biotinylated detection antibodies: anti-IL-17, anti-22 and anti-IL-21 (R&D systems), anti-IFN-γ, anti-IL-4 and anti-IL-10 (BD Bioscience).
Statistical analysis
Statistical analysis was performed using Student's t-test for most of the experiments. For colitis experiments, Man–Whitney test and analysis of variance test were used for comparison of disease score and weight loss. P values <0.05 were considered statistically significant.
Additional information
How to cite this article: Ouyang, X. et al. Transcription factor IRF8 directs a silencing programme for TH17-cell differentiation. Nat. Commun. 2:314 doi: 10.1038/ncomms1311 (2011).
References
Bettelli, E., Korn, T. & Kuchroo, V. K. Th17: the third member of the effector T cell trilogy. Curr. Opin. Immunol. 19, 652–657 (2007).
Weaver, C. T., Harrington, L. E., Mangan, P. R., Gavrieli, M. & Murphy, K. M. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity 24, 677–688 (2006).
Zheng, Y. et al. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007).
Liang, S. C. et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203, 2271–2279 (2006).
Veldhoen, M., Hocking, R. J., Atkins, C. J., Locksley, R. M. & Stockinger, B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006).
Korn, T. et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature 448, 484–487 (2007).
Nurieva, R. et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483 (2007).
Zhou, L. et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8, 967–974 (2007).
Mangan, P. R. et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441, 231–234 (2006).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Ivanov, I I et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
McGeachy, M. J. et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10, 314–324 (2009).
Yang, L. et al. IL-21 and TGF-beta are required for differentiation of human T(H)17 cells. Nature 454, 350–352 (2008).
Harrington, L. E. et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 (2005).
Stumhofer, J. S. et al. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat. Immunol. 7, 937–945 (2006).
Korn, T., Bettelli, E., Oukka, M. & Kuchroo, V. K. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27, 485–517 (2009).
Lee, C. H. et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 203, 63–72 (2006).
Zhao, J. et al. IRF-8/interferon (IFN) consensus sequence-binding protein is involved in Toll-like receptor (TLR) signaling and contributes to the cross-talk between TLR and IFN-gamma signaling pathways. J. Biol. Chem. 281, 10073–10080 (2006).
Tsujimura, H., Tamura, T. & Ozato, K. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J. Immunol. 170, 1131–1135 (2003).
Driggers, P. H. et al. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc. Natl Acad. Sci. USA 87, 3743–3747 (1990).
Barber, S. A., Fultz, M. J., Salkowski, C. A. & Vogel, S. N. Differential expression of interferon regulatory factor 1 (IRF-1), IRF-2, and interferon consensus sequence binding protein genes in lipopolysaccharide (LPS)-responsive and LPS-hyporesponsive macrophages. Infect. Immun. 63, 601–608 (1995).
Wang, H. & Morse, H. C. III IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol. Res. 43, 109–117 (2009).
Tamura, T., Nagamura-Inoue, T., Shmeltzer, Z., Kuwata, T. & Ozato, K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 13, 155–165 (2000).
Gabriele, L. & Ozato, K. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 18, 503–510 (2007).
Tamura, T., Yanai, H., Savitsky, D. & Taniguchi, T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584 (2008).
Ichiyama, K. et al. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J. Biol. Chem. 283, 17003–17008 (2008).
Zhang, F., Meng, G. & Strober, W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9, 1297–1306 (2008).
Akimzhanov, A. M., Yang, X. O. & Dong, C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 282, 5969–5972 (2007).
Zhang, Z., Zheng, M., Bindas, J., Schwarzenberger, P. & Kolls, J. K. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm. Bowel Dis. 12, 382–388 (2006).
Schraml, B. U. et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409 (2009).
Ozato, K., Tailor, P. & Kubota, T. The interferon regulatory factor family in host defense: mechanism of action. J. Biol. Chem. 282, 20065–20069 (2007).
Brustle, A. et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol. 8, 958–966 (2007).
Miossec, P., Korn, T. & Kuchroo, V. K. Interleukin-17 and type 17 helper T cells. N. Engl. J. Med. 361, 888–898 (2009).
Matsuzaki, G. & Umemura, M. Interleukin-17 as an effector molecule of innate and acquired immunity against infections. Microbiol. Immunol. 51, 1139–1147 (2007).
Ma, C. S. et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 205, 1551–1557 (2008).
Islander, U. et al. Superantigenic Staphylococcus aureus stimulates production of interleukin-17 from memory but not naive T cells. Infect. Immun. 78, 381–386 (2010).
Masumi, A., Tamaoki, S., Wang, I. M., Ozato, K. & Komuro, K. IRF-8/ICSBP and IRF-1 cooperatively stimulate mouse IL-12 promoter activity in macrophages. FEBS Lett. 531, 348–353 (2002).
Xiong, H. et al. Complex formation of the interferon (IFN) consensus sequence-binding protein with IRF-1 is essential for murine macrophage IFN-gamma-induced iNOS gene expression. J. Biol. Chem. 278, 2271–2277 (2003).
Kim, Y. M., Im, J. Y., Han, S. H., Kang, H. S. & Choi, I. IFN-gamma up-regulates IL-18 gene expression via IFN consensus sequence-binding protein and activator protein-1 elements in macrophages. J. Immunol. 165, 3198–3205 (2000).
Marecki, S., Riendeau, C. J., Liang, M. D. & Fenton, M. J. PU.1 and multiple IFN regulatory factor proteins synergize to mediate transcriptional activation of the human IL-1 beta gene. J. Immunol. 166, 6829–6838 (2001).
Xiong, H. et al. Ubiquitin-dependent degradation of interferon regulatory factor-8 mediated by Cbl down-regulates interleukin-12 expression. J. Biol. Chem. 280, 23531–23539 (2005).
Kong, H. J. et al. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J. Immunol. 179, 26–30 (2007).
Bettelli, E. & Kuchroo, V. K. IL-12- and IL-23-induced T helper cell subsets: birds of the same feather flock together. J. Exp. Med. 201, 169–171 (2005).
Zhou, L. et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240 (2008).
Leppkes, M. et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology 136, 257–267 (2009).
Seiderer, J. et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn′s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm. Bowel Dis. 14, 437–445 (2008).
O'Connor, W. Jr et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 10, 603–609 (2009).
De Jager, P. L. et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 41, 776–782 (2009).
Totsuka, T. et al. IL-7 Is essential for the development and the persistence of chronic colitis. J. Immunol. 178, 4737–4748 (2007).
Powrie, F., Leach, M. W., Mauze, S., Caddle, L. B. & Coffman, R. L. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 5, 1461–1471 (1993).
Acknowledgements
We thank C. Dong, A. Yoshimura, T. Taniguchi, O. Sakaguchi, M. Ono, H. Wang, J. Yang and G. Lu for providing reagents, technical supports and helpful suggestions. We are grateful to J. Unkeless, K. Honda and J. Blander for critical reading of the manuscript. This work was supported by the NIH P01 DK072201, a Crohn's and Colitis Foundation of America grant, a grant from the Eli and Edythe L. Broad Foundation, and by the intramural research programme of the NIH, National Institute of Allergy and Infectious Diseases and the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Author information
Authors and Affiliations
Contributions
X.O and H.X. initiated the project and designed the experiments. X.O. and R.Z. performed the majority of the experiments. J.Y., Q.L. and C.Z. performed experiments. L.Q. did histology analysis. J.L., H.N. performed ChIP assay experiments. M.S.S., C.-F.Q. and H.C.M provided IRF8 conditional KO mice. M.G. and K.O. generated various IRF8 mutant plasmids. J.C.H., S.A.L., and L.M. provided important reagents and critical discussion. X.O. and H.X. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1–S11 and Supplementary Table S1 (PDF 2389 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Ouyang, X., Zhang, R., Yang, J. et al. Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat Commun 2, 314 (2011). https://doi.org/10.1038/ncomms1311
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms1311
This article is cited by
-
MicroRNA expression profile of chicken cecum in different stages during Histomonas meleagridis infection
BMC Veterinary Research (2022)
-
Aryl hydrocarbon receptor activation drives polymorphonuclear myeloid-derived suppressor cell response and efficiently attenuates experimental Sjögren’s syndrome
Cellular & Molecular Immunology (2022)
-
T cell fate following Salmonella infection is determined by a STING-IRF1 signaling axis in mice
Communications Biology (2019)
-
Genome-wide interaction and pathway-based identification of key regulators in multiple myeloma
Communications Biology (2019)
-
Ankylosing spondylitis is associated with aberrant DNA methylation of IFN regulatory factor 8 gene promoter region
Clinical Rheumatology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.