Abstract
Oncogenic KRAS mutations introduce discrete amino acid substitutions that reduce intrinsic Ras GTPase activity and confer resistance to GTPase-activating proteins (GAPs). Here we discover a partial duplication of the switch 2 domain of K-Ras encoding a tandem repeat of amino acids G60_A66dup in a child with an atypical myeloproliferative neoplasm. K-Ras proteins containing this tandem duplication or a similar five amino acid E62_A66dup mutation identified in lung and colon cancers transform the growth of primary myeloid progenitors and of Ba/F3 cells. Recombinant K-RasG60_A66dup and K-RasE62_A66dup proteins display reduced intrinsic GTP hydrolysis rates, accumulate in the GTP-bound conformation and are resistant to GAP-mediated GTP hydrolysis. Remarkably, K-Ras proteins with switch 2 insertions are impaired for PI3 kinase binding and Akt activation, and are hypersensitive to MEK inhibition. These studies illuminate a new class of oncogenic KRAS mutations and reveal unexpected plasticity in oncogenic Ras proteins that has diagnostic and therapeutic implications.
Similar content being viewed by others
Introduction
Ras proteins are signal switch molecules that cycle between active GTP-bound and inactive GDP-bound conformations (Ras-GTP and Ras-GDP)1,2,3,4. Extracellular stimuli activate guanine nucleotide exchange factors, which enhance nucleotide exchange on Ras and thereby increase Ras-GTP levels2,3,4. GTP binding stabilizes the switch 1 and switch 2 domains of Ras and allows them to interact productively with downstream effectors including Raf and phosphatidylinositol 3-kinase (PI3K)2,3,4. Ras-GTP is hydrolysed to Ras-GDP through an intrinsic GTPase activity, which is augmented thousands of fold by GTPase-activating proteins (GAPs)2,3. Thus, the competing activities of guanine nucleotide exchange factors and GAPs regulate Ras output in vivo.
KRAS codons 12, 13 and 61 are the most common foci of dominant oncogenic mutations in human cancer4,5. Substitutions in these residues result in constitutively elevated levels of Ras-GTP due to reduced intrinsic GTP hydrolysis and resistance to GAPs2,3,4. Furthermore, germline KRAS mutations encoding gain-of-function proteins that are less activated biochemically than oncogenic K-Ras cause some cases of Noonan syndrome4,6,7. Taken together with the highly conserved nature of the Ras/GAP switch, the amino acid substitutions identified in cancer and Noonan syndrome suggest that a limited spectrum of mutations are capable of causing disease by constitutively activating Ras output. Here we describe insertion mutations within the switch 2 domain of KRAS that potently inhibit K-Ras intrinsic and GAP-mediated GTP hydrolysis. These mutations result in elevated MAPK signalling, but are defective in stimulating PI3K activity. Accordingly, cells transformed by these mutations show heightened sensitivity to growth inhibition by MEK inhibitors suggesting a therapeutic potential for the treatment of disease harbouring these or similar mutations.
Results
Identification of a KRAS insertion mutation in paediatric MPN
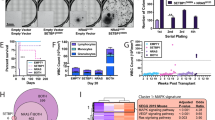
Juvenile myelomonocytic leukaemia (JMML) is an aggressive myeloproliferative neoplasm (MPN) characterized by driver Ras pathway mutations in 85% of cases, including known oncogenic KRAS and NRAS substitutions8. We discovered a partial duplication of the switch 2 domain of K-Ras in a 3-year-old boy with splenomegaly, pancytopenia and absolute monocytosis with abnormal cellular morphology (Fig. 1a) that persisted over 2.5 years of clinical observation (Supplementary Table 1). After multiple bone marrow examinations were not diagnostic for haematologic malignancy, blood leukocyte DNA was tested for mutations in the JMML genes NRAS, KRAS, PTPN11 and CBL. This analysis unexpectedly revealed a heterozygous partial KRAS duplication (39% allele frequency; c.178_198dup) resulting in the repetition of seven amino acids within the switch 2 domain (Fig. 1b,c) that was absent in a paired DNA specimen from his buccal mucosa. Database queries failed to uncover the same 21 nucleotide insertion in other tumour specimens; however, two lung adenocarcinomas and a colorectal cancer annotated in the COSMIC database contained a tandem duplication inserting five amino acids into K-Ras at the same location in the switch 2 domain (c.184_198dup; Fig. 1c). Immunoblot analysis of a protein lysate prepared from the patient’s blood mononuclear cells revealed a band with reduced electrophoretic mobility compared with the majority of total Ras protein (Fig. 1d). This band was not detected in normal leukocytes, and was of equivalent intensity to the normally migrating K-Ras species when probed with an antibody specific for K-Ras (Fig. 1d).
(a) Wright-Giemsa stained blood smear from the index patient shows a dyspoietic monocyte. (b) 454Jr pyrosequencing consensus reads showing 21-nucleotide tandem duplication of codons 60–66 of K-Ras. (c) K-Ras domain structure with amino acid numbers for the P-loop (PL), Switch 1 (SWI), Switch 2 (SWII) and hypervariable (HV) domains are shown. Blow-up of SWII depicts predicted individual amino acids of wild type, c.184_198dup, and c.178_198dup showing duplicated amino acids in red. (d) Western blot analysis of total Ras (lanes 1–3) and K-Ras (lanes 4 and 5) levels in the blood leukocytes of the index patient (1 and 4) and a normal individual (2 and 5). Black arrowhead indicates a higher molecular weight K-Ras protein in the patient sample. (e) CFU-GM colony formation by mouse fetal liver cells expressing GFP-tagged wild-type K-Ras (WT), K-RasG12D (D12), K-RasE62_A66dup (5AA), or K-RasG60_A66dup (7AA). Colony number is normalized to highest value for each genotype. Representative data from three independent experiments depicting the average±s.d. of technical replicates is shown. (f) Representative images of CFU-GM colonies from indicated genotypes grown at 0.1 ng ml−1 of GM-CSF. Scale bars, a, 10 μm; f, 100 μm.
KRAS duplication mutations transform myeloid progenitors
A hypersensitive pattern of colony-forming unit granulocyte macrophage (CFU-GM) progenitor formation in response to colony-stimulating factor (GM-CSF) is a cellular hallmark of JMML8,9. To ask whether K-Ras insertion mutant proteins can transform hematopoietic progenitor colony growth, we infected mouse fetal liver cells with vectors encoding wild-type (WT) or mutant K-Ras proteins (K-RasG60_A66dup, K-RasE62_A66dup or oncogenic K-RasG12D) fused to an amino-terminal green fluorescent protein (GFP) tag10. GFP-positive cells were isolated by cell sorting and plated in methylcellulose medium containing a range of GM-CSF concentrations. As expected7,11, K-RasG12D induced cytokine-independent CFU-GM colony formation and a hypersensitive pattern of growth characterized by increased numbers of abnormally large colonies at low GM-CSF concentrations (Fig. 1e). Expressing either K-Ras insertion also sensitized myeloid progenitors to GM-CSF, but had less pronounced effects on colony size (Fig. 1f).
KRAS insertion mutants disrupt the RAS–GAP interaction site
We next examined published crystal structures to model potential effects of switch 2 insertions on the following: (1) the positions of critical residues involved in intrinsic catalysis such as Glutamine 61 (Q61); (2) the Ras/GAP interface12; and (3) interactions with Raf13 and PI3K14. Due to the rigidity of the α2 helix at the C-terminal end of switch 2, inserting five or seven amino acids is expected to result in mostly local changes in protein structures. The inserted sequence is predicted to extend the N terminus of the α2 helix, yet there is uncertainty regarding the exact placement. To identify potential effects on Q61 and known protein–protein interactions, we generated 20 in silico models representing the possible orientations of the duplicated amino acids (Fig. 2a, Supplementary Fig. 1). Regardless of the specific placement, the extended structures are expected to overlap and disrupt the interaction of Ras with the α6 helix of GAP in the Ras–GAP complex (Fig. 2b), as well as possibly interfering with the location or orientation of Ras residue Q61 (Fig. 2a). While the majority of possible insertion positions do not provide a significant steric impediment to Ras–PI3K interactions, the potential exists for disrupting specific interaction between residues of switch 2 and PI3K (Fig. 2c). By contrast, existing structural data suggest little or no effect on the interaction between Ras and Raf, which involves only the switch 1 domain of Ras (Fig. 2d). Together this analysis indicates that tandem duplications in switch 2 likely favour the GTP conformation of Ras due to reduced interactions with GAP while preserving binding to Raf.
(a) Aggregate ribbon model depicting Ras (green, α2 helix in black) in its GTP-bound state, and 20 different possible orientations of the duplicated amino acids of switch 2 (rainbow). The inset shows predicted conformations of Q61; in most cases, Q61 is disordered, consistent with an active state switch 2 and reduced rate of intrinsic hydrolysis13. (b) Structural model from panel (a) superimposed into the co-crystal of the Ras/Ras–GAP (space filling) complex (PDB:1WQ1) showing steric interference at the binding interface; primary conflict is with α6 helix of GAP (inset). (c) Ras switch 2 mutant model in the context of Ras/PI3K interaction (PDB:1HE8). The extended helix does not provide a major steric impediment to binding face, but may disrupt orientation of specific interaction residues. (d) The extended helix is not predicted to perturb Raf binding to switch 1 (PDB:4G0N).
GTPase activity is reduced in KRAS duplication mutants
To directly test these predictions, we produced N-terminal histidine fusions encoding amino acids 1–166 of K-RasG60_A66dup or K-RasE62_A66dup, and compared their biochemical properties with WT K-Ras and K-RasG12D (Supplementary Fig. 2). Remarkably, recombinant insertion mutant proteins accumulated in the active GTP conformation to a substantially greater degree than K-RasG12D (Fig. 3a,b), and exhibited severely impaired intrinsic GTPase activity (Fig. 3c). We assayed the ability of the GAP-related domain of p120 GAP to stimulate GTP hydrolysis. As expected15, the GAP-related domain markedly enhanced the GTPase activity of WT K-Ras, whereas K-RasG12D was resistant (Fig. 3d). Consistent with our structural modelling, both insertion mutant proteins were insensitive to GAP stimulation (Fig. 3d).
(a) Phosphorimaging of α-P32 nucleotides eluted from indicated recombinant wild-type K-Ras (WT), K-RasG12D (D12), K-RasE62_A66dup (5AA), or K-RasG60_A66dup (7AA) proteins and separated by thin layer chromatography. Labelling, elution, and electrophoresis were performed in duplicate. (b) Quantification of the data shown in panel a indicating the percentage of GTP-bound Ras after 2 hr incubation. (c) Intrinsic GTP hydrolysis of recombinant K-Ras proteins. Scintillation counting of phosphates liberated from K-Ras proteins loaded with γ-P32 GTP at indicated time points. Best fit linear regressions are shown. Data are average±s.d. of duplicate reactions and are representative of three independent experiments. (d) Scintillation counting of free phosphates released by K-Ras proteins that were loaded with γ-P32 GTP and incubated with the indicated concentrations of a recombinant peptide encoding the GAP domain of p120 GAP. Data are average±s.d. of duplicate reactions and representative of three independent experiments.
KRAS insertion mutations impair PI3K binding and activation
Expression of K-RasG12D and each tandem duplication mutant, but not WT K-Ras, transformed interleukin 3 (IL-3)-dependent Ba/F3 cells to cytokine-independent growth (Supplementary Fig. 3a). Ba/F3 cells expressing K-RasG12D, K-RasG60_A66dup or K-RasE62_A66dup had elevated levels of Ras-GTP after being deprived of serum overnight, which did not increase further on IL-3 stimulation (Supplementary Fig. 3b).
To assess how acute activation of K-Ras duplication mutants modulates effector pathway activation, we engineered tetracycline inducible GFP-K-Ras constructs and introduced them into Ba/F3 cells (Supplementary Fig. 4). Inducing the expression of either K-Ras duplication mutant or of K-RasG12D reproducibly resulted in elevated levels of pERK compared with WT K-Ras (Fig. 4a). Interestingly, similar levels of K-RasQ61L induced far greater levels of ERK phosphorylation than either duplication mutant or K-RasG12D (Fig. 4a). These results are consistent with proposed allosteric effects of codon 61 Ras mutations on the Ras–Raf complex, which result in further reduction of GTP turnover16. By contrast, K-RasG12D or K-RasQ61L expression increased pAkt levels similarly and to a much greater extent than either insertion mutant in Ba/F3 cells (Fig. 4a).
(a) Western blot (top) and quantification (bottom) of lysates prepared from Ba/F3 cells expressing wild-type K-Ras (WT), K-RasG12D (D12), K-RasQ61L (L61), K-RasE62_A66dup (5AA), or K-RasG60_A66dup (7AA) after a 6 h starve in media containing 0.5 μg ml−1 doxycycline, no IL-3 and 1% FBS. Quantification shows the average±s.e.m. of four independent experiments. (b) Representative western blots (top) and quantifications (bottom) of recombinant effector binding to His–K-Ras proteins loaded with the indicated nucleotide. Quantification shows the average±s.d. of two independent experiments. (c,d) Dose response curves of Ba/F3 cells transformed with various K-Ras proteins that were grown for 3 days in the presence of the indicated concentrations of either PD0325901 (PD-901) (c) or GDC-0941 (d). Data are mean±s.e.m. of three wells and are representative of three independent experiments. Trend lines represent best fit to a fixed slope sigmoidal dose response equation. The 95% CI of GI50 values are indicated. a.u., arbitrary unit; CI, confidence interval.
Together with prior structural modelling predictions, these biochemical data prompted us to directly assess the ability of WT and mutant K-Ras proteins to bind to effectors in vitro. As expected, His–K-Ras WT bound GST–Raf–Ras-binding domain (RBD) and FLAG-p110α/p85 peptides in a nucleotide dependent fashion (Fig. 4b). All three mutant Ras proteins associated with GST–Raf–RBD to a similar extent. Importantly, however, K-RasG60_A66dup and K-RasE62_A66dup had a markedly reduced interaction with FLAG-p110α. A control K-RasG12D,Y64G protein, which contains a second site substitution that abrogates the Ras–PI3K interaction17, also showed profoundly reduced binding to FLAG-p110α (Fig. 4b).
KRAS insertion mutations show altered pathway dependencies
The observation that K-RasG12D and switch 2 insertion mutant proteins are defective for PI3K binding and Akt activation suggested that this might alter effector pathway dependencies. To address this question, we exposed transformed Ba/F3 cells to either a potent and selective MEK inhibitor (PD0325901) or to a pan-PI3K inhibitor (GDC-0941). Consistent with the differences observed in signalling outputs and effector interactions, the cytokine-independent growth of Ba/F3 cells transformed with either switch 2 insertion was more sensitive to MEK inhibition, but somewhat less sensitive to PI3K inhibition, than cells expressing K-RasG12D (Fig. 4c,d). These differences were not explained by either altered K-Ras expression levels (Supplementary Fig. 5) or differential target sensitivity, as low nanomolar concentrations of drug inhibited ERK phosphorylation with equivalent efficacy in all cell lines (Supplementary Fig. 6).
Discussion
Mutations in Ras pathway genes are a molecular hallmark of JMML8, an aggressive cancer that contains, on average, less than one additional somatic mutation per genome18. Furthermore, myeloid progenitors from Nf1, Kras, Nras and Ptpn11 mutant mice exhibit GM-CSF hypersensitivity and these strains all develop a JMML-like MPN19,20,21,22. Thus, clonal outgrowth of hematopoietic cells harbouring a somatic 21 nucleotide in frame duplication within the KRAS switch 2 domain in a child with persistent haematologic abnormalities strongly implicated this mutation as driving aberrant growth in vivo. Database searches revealed a similar 15 nucleotide insertion in primary colon and lung cancers, tumour types with frequent somatic KRAS mutations (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/)23,24. K-RasG60_A66dup and K-RasE62_A66dup proteins transformed the growth of primary myeloid progenitors and Ba/F3 cells, supporting both KRAS switch 2 insertions as bona fide oncogenic drivers.
Like canonical Ras oncoproteins, recombinant K-RasG60_A66dup and K-RasE62_A66dup accumulate in the GTP conformation due to defective intrinsic GTPase activity and resistance to GAP. Importantly, however, both switch 2 duplication/insertion mutant proteins exhibit novel biochemical properties. In particular, K-RasG60_A66dup and K-RasE62_A66dup are the first oncogenic Ras proteins with strong selectivity for Raf versus PI3K binding. This is unexpected given the large body of data implicating activation of both Raf/MEK/ERK and PI3K/Akt signalling downstream of oncogenic K-Ras as essential in JMML and lung cancer pathogenesis. Mx1-Cre, KrasG12D and Mx1-Cre, Nf1flox/flox mice develop a JMML-like MPN that is remarkably responsive to MEK inhibition in vitro and in vivo19,20,25,26 and genetically inactivating ERK or p110α profoundly attenuates MPN in this strain27,28. The atypical and indolent clinical course of MPN in our index patient further supports an essential role of activated PI3K signalling in JMML pathogenesis. Similarly, pharmacologic and genetic data strongly implicate concurrent activation of Raf/MEK/ERK and PI3K in KRAS-mediated lung cancer initiation and maintenance29,30,31. The situation is more complex in colon cancer where frequent co-occurrence of KRAS and PIK3CA mutations and a lack of concordance between levels of PI3K pathway activation and oncogenic K-Ras suppression are observed32,33. We emphasize that K-RasG60_A66dup and K-RasE62_A66dup retain some PI3K binding and more potently transform the growth of myeloid progenitors than K-RasG12D,Y64G.
Although KRAS switch 2 insertion/duplication mutations are infrequent in cancer sequence databases, they were previously detected in seminal studies of transforming factors in mice and rat34,35. Distinct KRAS mutations are associated with differential response to EGFR inhibition in colon cancer36. Similarly, our data suggest that MEK inhibition might be particularly effective in cancers with switch 2 insertion/duplication mutations. In addition, the observation that the KRASG60_A66dup and KRASE62_A66dup alleles are oncogenic despite impaired PI3K binding and Akt activation suggests that deep and sustained biochemical suppression by PI3K pathway inhibitors will likely be essential for clinical efficacy in RAS mutant cancers. Finally, several small molecule inhibitors of Ras were recently described, some of which make important contacts with switch 2 including the α2 helix and residues duplicated in patient specimens (Supplementary Fig. 7)37,38,39. While such inhibitors are in early stages of development, insertions into the switch 2 domain that reduce drug binding while retaining oncogenic activity are a potential mechanism of acquired resistance in KRAS mutant cancers. Consistent with this idea, an acute leukaemia that emerged in mice transplanted with hematopoietic cells expressing K-RasG12D,Y64G acquired a de novo switch 2 insertion that restored full oncogenic activity11.
Methods
Mutation detection by pyrosequencing
All human samples were collected and analysed under the approval of UCSF’s committee on human research CHR # 10-04212. Genomic DNA was extracted on the Qiagen EZ1 and amplified by standard PCR using primers targeted to exons 8 and 9 of CBL (NM_005188.3), exons 2 and 3 of KRAS (NM_004985.3), exons 2 and 3 of NRAS (NM_002524.4) and exons 3, 4 and 13 of PTPN11 (NM_002834.3). Amplicons were sequenced on the Roche 454 GS Junior, and analysed with Roche Amplicon Variant Analysis software. The sensitivity of detecting a point mutation is 15% relative to that of the normal DNA sequence.
Detection of mutant Ras proteins in from human samples
Blood drawn into heparin vacutainers was diluted in Hanks’ balanced salt solution (Life technologies) and subjected to centrifugation over Ficoll-paque PLUS (GE healthcare). Recovered mononuclear cells were washed and pelleted in Hanks’ balanced salt solution. Approximately one million cells were lysed by intermittent vortexing in 100 μl cold lysis buffer containing 25 mM HEPES pH 7.5, 150 mM NaCl, 20 mM NaF, 2 mM Na3VO4, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS and EDTA-free protease inhibitor tablets (Pierce). Lysates were clarified by 20,000 × r.c.f. centrifugation, and combined 5:1 with 6 × SDS sample buffer (50% glycerol, 300 mM Tris, pH 6.8, 600 mM DTT, 12% SDS, 0.06% bromophenol blue) before heating to 95–100 °C for 5 min. Samples were separated on criterion TGX gels (Bio-Rad) and transferred to Imobilon-FL membrane (Millipore). Membranes were blocked for 1 h with Odyssey Blocking Buffer (Li-Cor) containing 0.1% Tween-20 before incubation with primary antibodies (Supplementary Table 2) for 3–4 h at room temperature. After primary incubation, membranes were washed four times for 5 min in tris-buffered saline (Bio-Rad, #170–6,435) containing 0.1% Tween-20. After washing, membranes were incubated with a 1:5,000 dilution of secondary florescent antibodies (Li-Cor, 926–68,071 and 926–32,210) in blocking buffer plus 0.01% SDS. After a 1 h incubation, membranes were washed as above and then scanned using a Li-Cor odyssey florescent scanner. Quantifications were performed using Image studio light (Li-Cor).
Cloning of GFP–K-Ras mutants
Kras Q61L, Kras 5AA and Kras 7AA were created by performing mutagenesis of a pEGFP–KRas WT plasmid by amplification with phosphorylated oligonucleotides Q61L_Forward 5′-ACAGCAGGTCTAGAGGAGTACAGT-3′, Q61L_Reverse 5′-GTCGAGAATATCCAAGAGACAGG-3′, 5AA_Forward 5′-TGCACTGTACTCCTCTTGACCTGC-3′, 5AA_Reverse 5′-GAGGAGTACAGTGCAATGAGGGACCAGTACATGAGAACTGG-3′, 7AA_Forward 5′-GGTCAAGAGGAGTACAGTGCAGGTCAAGAGGAGTACAGTGCAATG-3′, 7AA_Reverse 5′-TGCTGTGTCGAGAATATCCAAGAG-3′ using phusion DNA polymerase. DNA coding for the EGFP–KRas Q61L and EGFP–KRas insertion mutants was then sub cloned from pEGFP into pMIG puro using NcoI/SalI restriction sites. Doxycycline-induced GFP–KRAS WT and mutant constructs were generated by amplification of GFP–Kras sequence with phusion DNA polymerase using the following primers: 5′-GTACCCGGGGATCTGATCACTCGAGCCACCATGGTGAGCAAGGGCGAG-3′, 5′-GTATAATGTATACTTAATTAATCTAGATCACATAACTGTACACCTTG-3′. Following amplification, PCR products were introduced into a retroviral plasmid containing a Tet responsive element upstream of a PGK-driven Puromycin resistance cassette prepared by XhoI and XbaI digestion, via Gibson Assembly.
Retroviral production
Freshly split, 10 cm dishes of HEK 293 T cells were transfected with 10 μg of pMIG puro EGFP–Kras and 6 μg Eco–Pak packaging plasmid using calcium phosphate. Following overnight incubation, cells were switched into fresh media and allowed to produce virus for 48 h. Viral-containing media was centrifuged for 1 h in microcentrifuge tubes at 20,000 r.c.f. at 4 °C. After centrifugation, 60% of the media was removed and viral pellets were suspended in remaining volume before snap freezing and storage at −80 °C.
Fetal liver transduction and GM-CFU assays
All animal experiments conformed to national regulatory standards and were approved by the UCSF Committee on Animal Research protocol #AN091877-03B. Livers were surgically removed from E14–14.5 C57BL/6 mice and then dissociated by trituration in IMDM media containing 20% FBS with a 19.5 gage needle before cryopreservation in liquid nitrogen. Fetal liver cells were thawed into IMDM media, recovered by centrifugation and then RBC lysed in RBC lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 1 mM EDTA pH 7.3). After RBC lysis, fetal liver cells were suspended in stim media consisting of StemSpan (StemCell Technologies) containing 10 mM HEPES buffer pH 7.4, GlutaMAX (Life Technologies), penicillin/streptomycin, 15% FBS (HyClone), 10 ng ml−1 IL-3, 10 ng ml−1 SCF, 10 ng ml−1 IL-6 (PeproTech) and 50 μM β-Mercaptoethanol, and passed through a 70 micron mesh filter. After overnight culture, fetal liver cells were transduced with indicated retroviruses by ‘spinfection’ in the presence of concentrated viral supernatant supplemented with 5 μg ml−1 polybrene for 2.5 h at 800 r.c.f. Following spinfection, cells were returned to stim media. Cells were cultured for an additional 2 days before sorting for GFP positivity using a FACS Aria III cell sorter. GFP-positive cells were sorted directly into MethoCult media (StemCell Technologies) supplemented with GlutaMAX and penicillin/streptomycin before addition of indicated concentrations of GM-CSF. Supplemented MethoCult media and 10,000 GFP-positive cells were then plated into 35 mm dishes and incubate for 7 days before counting of colonies.
Structure modelling
K-Ras proteins containing switch 2 duplications were first submitted to the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/)40 for structure predictions. The top 10 models showed a diverse set of potential conformations for the inserted sequences (Supplementary Fig. 1). Although it is difficult to predict the precise orientations of the insertions, the α2-helix (M67-R73) at the C-terminal end of switch 2 maintains helical conformation in each model. In addition, secondary structure prediction indicates that the amino acids immediately preceding the α2-helix (EETSA) have strong helix propensities. Therefore, the insertions are likely to extend the N terminus of the α2-helix, suggesting that the insertions should be more structured than the automatic server predicts. To refine the structure prediction, we used Modeller9v7 (https://salilab.org/modeller/)41 to create homology models with a published Ras structure (PDB:1WQ1) as the template. For K-RasG60-A66dup, rather than modelling just the insertion sequence (GQEEYSA), the entire 13 amino acid region (QEEYSAGQEEYSA) was modelled in the context of the whole molecule, but without using any structure information from the template. To investigate the potential structural impact of insertion on protein–protein interactions, 20 models were generated. These models all contain an elongated α2-helix and native-like conformations for the disordered regions. Modelling the structure of the 11 amino acid region (QEEYSAEEYSA) in K-RasG62-A66dup using the same approach yielded similar results.
Cloning of pET 28a bacterial expression constructs
K-Ras WT and mutant sequences were amplified with phusion DNA polymerase using the following primers: 5′-GGCCTGGTGCCGCGCGGCAGCCATATGACTGAGTATAAACTTGTGGTG-3′, 5′-GAGTGCGGCCGCAAGCTTGTCGACGGAGCTCCTAATGTTTTCGAATTTCTCGGA-3′. Following amplification, PCR products were introduced into pET 28a plasmid prepared by NdeI and EcoRI digestion, via Gibson Assembly. A pET28 human p120 GAP construct was prepared analogously using primers: 5′-GGCCTGGTGCCGCGCGGCAGCCATATGGAAAAAATCATGCCAG-3′, 5′-GAGTGCGGCCGCAAGCTTGTCGACGGAGCTCCTACCTGACATCATTGGTTTTTG-3′.
Bacterial expression and purification
Recombinant proteins were produced by transforming a BL21 CodonPlus (DE3) RIPL Escherichia coli strain with pET28 constructs. Lysate from 2 l cultures were purified using Ni-NTA resin (IMAC) and eluted in imidazole contacting buffer. Purified proteins were subjected to buffer exchange on desalting columns into 50 mM Tris pH 7.5, 200 mM NaCl, 10% glycerol and 5 mM β-mercaptoethanol. Proteins were quantified by Bradford assay.
Steady state levels of Ras nucleotide
For each reaction, 4 μg of His–K-Ras WT, G12D or insertion mutant protein was loaded with α32P-GTP by incubation in EDTA and nucleotide. Loading reactions were quenched by the addition of Excess buffer A (20 mM HEPES, 2 mM DTT, 2 mM) and incubated for 2 h at 37 °C. After incubations, K-Ras proteins were immunoprecipitated by the addition of 20 μl of anti-Ras–agarose slurry (Santa Cruz Biotechnology, sc-35 AC) followed by rocking agitation for 20 min at room temperature. Beads were washed four times with wash buffer (50 mM HEPES, 500 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, 0.005% SDS). Washed beads were suspended in elution buffer (2 mM EDTA, 2 mM DTT, 0.2% SDS, 0.5 mM GTP, 0.5 mM GDP) and heated to 70 °C for 20 min, cooled and then collected by centrifugation. Supernatants from the previous step were spotted on polyethylenimine-cellulose chromatography plates and run in 1 M LiCl. Following separation, plates were dried and exposed to phosphorimaging screens before acquisition using a storm 860 imager.
Basal and GAP stimulated GTP Hydrolysis
For each reaction, 4 μg of His–K-Ras WT, G12D or insertion mutant protein was loaded with γ32P-GTP as above. Loading reactions were quenched, and hydrolysis was initiated by the addition of excess pre-warmed buffer A (20 mM HEPES, 2 mM DTT, 2 mM). Reactions were then incubated at 37 °C and aliquots were removed at indicated time points and diluted into stop buffer (5 mM silicotungstate, 1 mM H2SO4). To assess levels of free 32P in stopped reactions, inorganic phosphate was chelated with molybdate and extracted into isobutene/toluene before quantification with a liquid scintillation counter. GAP-mediated hydrolysis was performed essentially as above except that reactions contained indicated amounts of p120 GAP and were run for 8 min at room temperature.
IL-3 withdrawal experiments
Ba/F3 cells, kindly provided by Neil Shah, were grown in RPMI 1640 media (Life Technologies) containing 10% FBS, GlutaMAX, penicillin/streptomycin, 0.5 mM sodium pyruvate, 50 μM β-Mercaptoethanol and 10 ng ml−1 IL-3. BaF3 cells were infected with pMIG puro EGFP-K-Ras constructs by overnight exposure to viral supernatant diluted 1:5 into normal growth media. Two days after infection cells were selected in normal growth media supplemented with 1 μg ml−1 puromycin for 4 days. Following selection, cells expressing each construct were collected by centrifugation at 600 × r.c.f., washed with growth media containing no IL-3, repelleted, and then resuspended in IL-3 lacking media at a ∼100,000 cells per ml. Cells were counted immediately (t=0) and then plated in triplicate wells of 12-well plates for subsequent counting at indicated time points.
Ras–Raf–RBD interactions
5,000,000 Ba/F3 cells were starved overnight in normal growth media containing 1% FBS and no IL-3. Cells were then stimulated or not with 5 ng ml−1 IL-3 for 30 min before collection by centrifugation. Collected cells were lysed in MLB buffer (10% glycerol, 25 mM HEPES pH 7.5, 150 mM NaCl, 25 mM NaF, 10 mM MgCl2, 1 mM Na3VO4, 1 mM EDTA, 1% IGEPAL CA-630, 0.25% sodium deoxycholate and protease inhibitor tablets). Lysates were clarified by centrifugation and then incubated with 20 μl of Raf–RBD agarose slurry (Milipore/upstate, 14–278) for 45 min with agitation at 4 °C. Following binding, beads were washed 3 × with MLB buffer and then boiled in 1 × SDS sample buffer before immunoblot analysis as above.
Generation of doxycycline inducible GFP–K-Ras expressing Ba/F3 cells
Ba/F3 cells were infected with murine RIEN retrovirus encoding the bicistronic transcript composed of the rtTA3 third generation reverse tetracycline-controlled transactivator, an internal ribosomal entry site, and the ecotropic murine retroviral receptor mCAT-1. This virus also expresses a neomycin resistance gene from a PGK promoter. Ba/F3 cells infected with this virus were then selected in normal growth media supplemented with 0.5 mg ml−1 G418 for 1 week before infection with TtIP virus for the various GFP–K-Ras constructs. After the TtIP infection Ba/F3 cells were grown in selection media containing G418 and Puromycin at 0.8 μg ml−1.
Assessment of acute effects of GFP-K-Ras expression on Ba/F3 cells
Stable cell lines generated as described above were counted, pelleted and resuspended in RPMI media containing no IL-3 and 1% FBS at 1.6 million cells per ml. Cells were divided into two six-well wells and doxycycline was added to one well at a final concentration of 0.5 μg ml−1. Cells were cultured for 6 h before 1.4 ml worth of cells was collected by centrifugation and snap freezing. Protein lysates were prepared and analysed as above. Uncropped images of immune blots from Fig. 4 are provided in Supplementary Fig. 8.
Recombinant effector interactions
5 μg of His–K-Ras WT, or mutant protein was loaded with GTP or GDP as indicated. Nucleotide loaded His–K-Ras proteins were diluted into cold binding buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 25 mM imidazole, 5 mM MgCl2, 0.5 mM DTT 0.1% Triton X-100) then incubated with TALON magnetic beads (Clonetech) with agitation at 4 °C for 20 min. Flag-p110α·p85 (Sigma), and GST–Raf-RBD (EMD-Millipore) were added to K-Ras beads at final concentrations of 1.2 ng μl−1 and 2.4 ng μl−1, respectively, and incubated for an additional 45 min at 4 °C. Following binding, beads were washed four times with cold binding buffer and then heated in 1 × SDS sample buffer. Bound proteins were analysed by immunoblot as above.
Drugs
GDC-0941 was provided by Genentech, Inc. (South San Francisco, CA). PD0325901 was synthesized by Shanghai Chempartner. All drugs were diluted in DMSO for in vitro studies.
Compound sensitivity assays
IL-3-independent Ba/F3 cells expressing K-Ras mutants were obtained from IL-3 withdrawal experiments. Cells were plated in triplicate wells of a 96-well plate in growth media without IL-3 and indicated concentrations of PD0325901, GDC-0491 or DMSO. Cells were cultured for 3 days and then cell viability was assessed using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to manufacturer’s recommended protocol.
Additional information
How to cite this article: White, Y. et al. KRAS insertion mutations are oncogenic and exhibit distinct functional properties. Nat. Commun. 7:10647 doi: 10.1038/ncomms10647 (2016).
References
Vetter, I. R. & Wittinghofer, A. The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001).
Donovan, S., Shannon, K. M. & Bollag, G. GTPase activating poteins: critical regulators of intracellular signaling. Biochim Biophys Acta 1602, 23–45 2002.
Bos, J. L., Rehmann, H. & Wittinghofer, A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 (2007).
Schubbert, S., Shannon, K. & Bollag, G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308 (2007).
Malumbres, M. & Barbacid, M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3, 459–465 (2003).
Schubbert, S. et al. Biochemical and functional characterization of germ line KRAS mutations. Mol. Cell. Biol. 27, 7765–7770 (2007).
Schubbert, S. et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38, 331–336 (2006).
Loh, M. L. Recent advances in the pathogenesis and treatment of juvenile myelomonocytic leukaemia. Br. J. Haematol. 152, 677–687 (2011).
Emanuel, P. D., Bates, L. J., Castleberry, R. P., Gualtieri, R. J. & Zuckerman, K. S. Seletive hypersensitivity to granulocyte-macrophage colony stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood 77, 925–929 (1991).
Schubbert, S. et al. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood 106, 311–317 (2005).
Shieh, A. et al. Defective K-Ras oncoproteins overcome impaired effector activation to initiate leukemia in vivo. Blood 121, 4884–4893 (2013).
Scheffzek, K. et al. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997).
Fetics, S. K. et al. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure. 23, 505–516 (2015).
Pacold, M. E. et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–943 (2000).
Bollag, G. et al. Biochemical characterization of a novel KRAS insertional mutation from a human leukemia. J. Biol. Chem. 273, 32491–32494 (1996).
Buhrman, G., Kumar, V. S., Cirit, M., Haugh, J. M. & Mattos, C. Allosteric modulation of Ras-GTP is linked to signal transduction through RAF kinase. J. Biol. Chem. 286, 3323–3331 (2011).
Rodriguez-Viciana, P. et al. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89, 457–467 (1997).
Sakaguchi, H. et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat. Genet. 45, 937–941 (2013).
Braun, B. S. et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc. Natl Acad. Sci. USA 101, 597–602 (2004).
Le, D. T. et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood 103, 4243–4250 (2004).
Li, Q. et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 117, 2022–2032 (2011).
Araki, T. et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 10, 849–857 (2004).
Schmid, K. et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin. Cancer Res. 15, 4554–4560 (2009).
Wojcik, P. et al. KRAS mutation profile in colorectal carcinoma and novel mutation–internal tandem duplication in KRAS. Pol. J. Pathol. 59, 93–96 (2008).
Chang, T. et al. Sustained MEK inhibition abrogates myeloproliferative disease in Nf1 mutant mice. J. Clin. Invest. 123, 335–339 (2013).
Lyubynska, N. et al. A MEK inhibitor abrogates myeloproliferative disease in Kras mutant mice. Sci. Transl. Med. 3, 76ra27 (2011).
Staser, K. et al. Normal hematopoiesis and neurofibromin-deficient myeloproliferative disease require Erk. J. Clin. Invest. 123, 329–334 (2013).
Gritsman, K. et al. Hematopoiesis and RAS-driven myeloid leukemia differentially require PI3K isoform p110alpha. J. Clin. Invest. 124, 1794–1809 (2014).
Gupta, S. et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell 129, 957–968 (2007).
Castellano, E. et al. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell 24, 617–630 (2013).
Blasco, R. B. et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663 (2011).
Ebi, H. et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J. Clin. Invest. 121, 4311–4321 (2011).
Cancer Genome . Atlas, N., Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Higinbotham, K. G., Rice, J. M., Buzard, G. S. & Perantoni, A. O. Activation of the K-ras gene by insertion mutations in chemically induced rat renal mesenchymal tumors. Oncogene 9, 2455–2459 (1994).
Wiest, J. S., Burnett, V. L., Anderson, M. W. & Reynolds, S. H. A novel mechanism of in vivo ras gene activation involving tandem duplication of coding sequences. Oncogene 9, 2449–2454 (1994).
Chen, J., Ye, Y., Sun, H. & Shi, G. Association between KRAS codon 13 mutations and clinical response to anti-EGFR treatment in patients with metastatic colorectal cancer: results from a meta-analysis. Cancer Chemother. Pharmacol. 71, 265–272 (2013).
Sun, Q. et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. 51, 6140–6143 (2012).
Maurer, T. et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl Acad. Sci. USA 109, 5299–5304 (2012).
Ostrem, J. M., Peters, U., Sos, M. L., Wells, J. A. & Shokat, K. M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Acknowledgements
We are grateful to Pankaj Vats and Arul Chinnaiyan, (University of Michigan) for extensive bioinformatic analysis of cancer databases, to Deepak Sampath (Genentech, Inc.) for providing the GDC-0941 used in this work, and to Frank McCormick for helpful suggestions. This work was supported by NIH grant R37CA72614 (to K.S.), by a Damon Runyon Cancer Research Foundation Fellowship DRG-2149-13 (to A.F.), and by a Howard Hughes Medical Institute Medical Student Research Fellowship (to Y.W). K.S. is an American Cancer Society Research Professor.
Author information
Authors and Affiliations
Contributions
Y.W. assisted with experimental design, performed research studies, analyzed data; A.B. & D.D. identified the index patient, performed diagnostic testing, contributed clinical data, and edited the manuscript; J.V.Z. & M.L. performed and analyzed mutation testing on the index patient and edited the manuscript; A.I. performed research studies; G.B. designed experiments, and edited the manuscript; C.Z. performed structural modelling, and wrote the manuscript; H.C. generated recombinant proteins; K.S. designed experiments, reviewed the data, and wrote the manuscript; A.J.F. designed experiments, performed research studies, analyzed data; and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
G.B., C.Z., and H.C. are full time employees of Plexxikon Inc. The remaining authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-8 and Supplementary Tables 1-2. (PDF 1011 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
White, Y., Bagchi, A., Van Ziffle, J. et al. KRAS insertion mutations are oncogenic and exhibit distinct functional properties. Nat Commun 7, 10647 (2016). https://doi.org/10.1038/ncomms10647
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms10647
This article is cited by
-
Excited-state observation of active K-Ras reveals differential structural dynamics of wild-type versus oncogenic G12D and G12C mutants
Nature Structural & Molecular Biology (2023)
-
Functional characterisation of a novel class of in-frame insertion variants of KRAS and HRAS
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.