Abstract
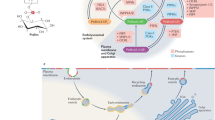
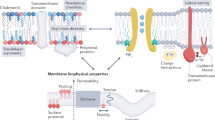
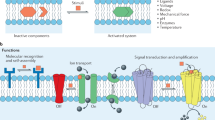
Membrane lipids function as structural molecules, reservoirs for second messengers, membrane platforms that scaffold protein assembly and regulators of enzymes and ion channels. Such diverse lipid functions contribute substantially to cellular mechanisms for fine-tuning membrane-signaling events. Meaningful coordination of these events requires exquisite spatial and temporal control of lipid metabolism and organization, and reliable mechanisms for specifically coupling these parameters to dedicated physiological processes. Recent studies suggest such integration is linked to the action of phosphatidylinositol transfer proteins that operate at the interface of the metabolism, trafficking and organization of specific lipids.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
van Meer, G. Cellular lipidomics. EMBO J. 24, 3159–3165 (2005).
Berridge, M.J. & Irvine, R.F. Inositol phosphates and cell signalling. Nature 341, 197–205 (1989).
Majerus, P.W. Inositol phosphate biochemistry. Annu. Rev. Biochem. 61, 225–250 (1992).
Rhee, S.G. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 70, 281–312 (2001).
Nishizuka, Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 9, 484–496 (1995).
Hurley, J.H. & Meyer, T. Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 13, 146–152 (2001).
Lemmon, M.A. Phosphoinositide recognition domains. Traffic 4, 201–213 (2003).
Shaw, G. The pleckstrin homology domain: an intriguing multifunctional protein module. Bioessays 18, 35–46 (1996).
Suh, B.C. & Hille, B. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr. Opin. Neurobiol. 15, 370–378 (2005).
Simonsen, A., Wurmser, A.E., Emr, S.D. & Stenmark, H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13, 485–492 (2001).
Martin, T.F. PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13, 493–499 (2001).
Wenk, M.R. & De Camilli, P. Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals. Proc. Natl. Acad. Sci. USA 101, 8262–8269 (2004).
Defacque, H. et al. Phosphoinositides regulate membrane-dependent actin assembly by latex bead phagosomes. Mol. Biol. Cell 13, 1190–1202 (2002).
Janmey, P.A. & Lindberg, U. Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cell Biol. 5, 658–666 (2004).
York, J.D., Odom, A.R., Murphy, R., Ives, E.B. & Wente, S.R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285, 96–100 (1999).
Odom, A.R., Stahlberg, A., Wente, S.R. & York, J.D. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287, 2026–2029 (2000).
Kronke, M. Biophysics of ceramide signaling: interaction with proteins and phase transition of membranes. Chem. Phys. Lipids 101, 109–121 (1999).
Pettus, B.J., Chalfant, C.E. & Hannun, Y.A. Ceramide in apoptosis: an overview and current perspectives. Biochim. Biophys. Acta 1585, 114–125 (2002).
Pettus, B.J., Chalfant, C.E. & Hannun, Y.A. Sphingolipids in inflammation: roles and implications. Curr. Mol. Med. 4, 405–418 (2004).
Spiegel, S. & Milstien, S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4, 397–407 (2003).
Stahelin, R.V., Rafter, J.D., Das, S. & Cho, W. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVa cytosolic phospholipase A2. J. Biol. Chem. 278, 12452–12460 (2003).
Iwamoto, K. et al. Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells 9, 891–903 (2004).
Moolenaar, W.H. Lysophosphatidic acid signalling. Curr. Opin. Cell Biol. 7, 203–210 (1995).
Exton, J.H. Signaling through phosphatidylcholine breakdown. J. Biol. Chem. 265, 1–4 (1990).
Singer, W.D., Brown, H.A. & Sternweis, P.C. Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D. Annu. Rev. Biochem. 66, 475–509 (1997).
Sciorra, V.A. et al. Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J. Cell Biol. 159, 1039–1049 (2002).
Liscovitch, M. & Cantley, L.C. Signal transduction and membrane traffic: the PITP/phosphoinositide connection. Cell 81, 659–662 (1995).
Hinchliffe, K.A., Ciruela, A. & Irvine, R.F. PIPkins1, their substrates and their products: new functions for old enzymes. Biochim. Biophys. Acta 1436, 87–104 (1998).
Cleves, A., McGee, T. & Bankaitis, V. Phospholipid transfer proteins: a biological debut. Trends Cell Biol. 1, 30–34 (1991).
Schwille, P., Haupts, U., Maiti, S. & Webb, W.W. Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophys. J. 77, 2251–2265 (1999).
Furst, W. & Sandhoff, K. Activator proteins and topology of lysosomal sphingolipid catabolism. Biochim. Biophys. Acta 1126, 1–16 (1992).
Wirtz, K.W. Phospholipid transfer proteins revisited. Biochem. J. 324, 353–360 (1997).
Hsuan, J. & Cockcroft, S. The PITP family of phosphatidylinositol transfer proteins. Genome Biol. 2 REVIEWS3011 (2001).
Phillips, S.E. et al. The diverse biological functions of phosphatidylinositol transfer proteins in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 41, 21–49 (2006).
Sha, B., Phillips, S.E., Bankaitis, V.A. & Luo, M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510 (1998).
Phillips, S.E. et al. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell 4, 187–197 (1999).
Yoder, M.D. et al. Structure of a multifunctional protein. Mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J. Biol. Chem. 276, 9246–9252 (2001).
Schouten, A. et al. Structure of apo-phosphatidylinositol transfer protein alpha provides insight into membrane association. EMBO J. 21, 2117–2121 (2002).
Tilley, S.J. et al. Structure-function analysis of human [corrected] phosphatidylinositol transfer protein alpha bound to phosphatidylinositol. Structure 12, 317–326 (2004).
Hanada, K. et al. Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809 (2003).
Roderick, S.L. et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 9, 507–511 (2002).
Li, X. et al. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157, 63–77 (2002).
Im, Y.J., Raychaudhuri, S., Prinz, W.A. & Hurley, J.H. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437, 154–158 (2005).
Kostenko, E.V., Mahon, G.M., Cheng, L. & Whitehead, I.P. The Sec14 homology domain regulates the cellular distribution and transforming activity of the Rho-specific guanine nucleotide exchange factor Dbs. J. Biol. Chem. 280, 2807–2817 (2005).
Sirokmany, G. et al. Sec14 homology domain targets p50RhoGAP to endosomes and provides a link between Rab and Rho GTPases. J. Biol. Chem. 281, 6096–6105 (2006).
Gu, M., Warshawsky, I. & Majerus, P.W. Cloning and expression of a cytosolic megakaryocyte protein-tyrosine-phosphatase with sequence homology to retinaldehyde-binding protein and yeast SEC14p. Proc. Natl. Acad. Sci. USA 89, 2980–2984 (1992).
Huynh, H. et al. Homotypic secretory vesicle fusion induced by the protein tyrosine phosphatase MEG2 depends on polyphosphoinositides in T cells. J. Immunol. 171, 6661–6671 (2003).
Whitehead, I.P., Campbell, S., Rossman, K.L. & Der, C.J. Dbl family proteins. Biochim. Biophys. Acta 1332, F1–23 (1997).
Rossman, K.L. et al. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 21, 1315–1326 (2002).
Cheng, L. et al. RhoGEF specificity mutants implicate RhoA as a target for Dbs transforming activity. Mol. Cell. Biol. 22, 6895–6905 (2002).
D'Angelo, I., Welti, S., Bonneau, F. & Scheffzek, K. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 7, 174–179 (2006).
Kearns, M.A. et al. Novel developmentally regulated phosphoinositide binding proteins from soybean whose expression bypasses the requirement for an essential phosphatidylinositol transfer protein in yeast. EMBO J. 17, 4004–4017 (1998).
Monks, D.E., Aghoram, K., Courtney, P.D., DeWald, D.B. & Dewey, R.E. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell 13, 1205–1219 (2001).
Min, K.C., Kovall, R.A. & Hendrickson, W.A. Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: implications for ataxia with vitamin E deficiency. Proc. Natl. Acad. Sci. USA 100, 14713–14718 (2003).
Stocker, A., Tomizaki, T., Schulze-Briese, C. & Baumann, U. Crystal structure of the human supernatant protein factor. Structure 10, 1533–1540 (2002).
Panagabko, C. et al. Ligand specificity in the CRAL-TRIO protein family. Biochemistry 42, 6467–6474 (2003).
Rogers, D.P. & Bankaitis, V.A. Phospholipid transfer proteins and physiological functions. Int. Rev. Cytol. 197, 35–81 (2000).
Hay, J.C. & Martin, T.F. Phosphatidylinositol transfer protein required for ATP-dependent priming of Ca(2+)-activated secretion. Nature 366, 572–575 (1993).
Ohashi, M. et al. A role for phosphatidylinositol transfer protein in secretory vesicle formation. Nature 377, 544–547 (1995).
Cunningham, E. et al. The yeast and mammalian isoforms of phosphatidylinositol transfer protein can all restore phospholipase C-mediated inositol lipid signaling in cytosol-depleted RBL-2H3 and HL-60 cells. Proc. Natl. Acad. Sci. USA 93, 6589–6593 (1996).
Simon, J.P. et al. An essential role for the phosphatidylinositol transfer protein in the scission of coatomer-coated vesicles from the trans-Golgi network. Proc. Natl. Acad. Sci. USA 95, 11181–11186 (1998).
Jones, S.M., Alb, J.G., Jr., Phillips, S.E., Bankaitis, V.A. & Howell, K.E. A phosphatidylinositol 3-kinase and phosphatidylinositol transfer protein act synergistically in formation of constitutive transport vesicles from the trans-Golgi network. J. Biol. Chem. 273, 10349–10354 (1998).
Cleves, A.E. et al. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 64, 789–800 (1991).
Bankaitis, V.A., Aitken, J.R., Cleves, A.E. & Dowhan, W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347, 561–562 (1990).
Guo, S., Stolz, L.E., Lemrow, S.M. & York, J.D. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274, 12990–12995 (1999).
Hama, H., Schnieders, E.A., Thorner, J., Takemoto, J.Y. & DeWald, D.B. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 34294–34300 (1999).
Rivas, M.P. et al. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol. Biol. Cell 10, 2235–2250 (1999).
Li, X. et al. Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol. Biol. Cell 11, 1989–2005 (2000).
Schnabl, M. et al. Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur. J. Biochem. 270, 3133–3145 (2003).
Routt, S.M. et al. Nonclassical PITPs activate PLD via the Stt4p PtdIns-4-kinase and modulate function of late stages of exocytosis in vegetative yeast. Traffic 6, 1157–1172 (2005).
Sreenivas, A., Patton-Vogt, J.L., Bruno, V., Griac, P. & Henry, S.A. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J. Biol. Chem. 273, 16635–16638 (1998).
Xie, Z. et al. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. USA 95, 12346–12351 (1998).
Xie, Z., Fang, M. & Bankaitis, V.A. Evidence for an intrinsic toxicity of phosphatidylcholine to Sec14p-dependent protein transport from the yeast Golgi complex. Mol. Biol. Cell 12, 1117–1129 (2001).
Wu, W.I., Routt, S., Bankaitis, V.A. & Voelker, D.R. A new gene involved in the transport-dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the phosphatidylinositol/phosphatidylcholine transfer protein, SEC14. J. Biol. Chem. 275, 14446–14456 (2000).
Wu, W.I. & Voelker, D.R. Reconstitution of phosphatidylserine transport from chemically defined donor membranes to phosphatidylserine decarboxylase 2 implicates specific lipid domains in the process. J. Biol. Chem. 279, 6635–6642 (2004).
Wu, W.I. & Voelker, D.R. Biochemistry and genetics of interorganelle aminoglycerophospholipid transport. Semin. Cell Dev. Biol. 13, 185–195 (2002).
Rudge, S.A. et al. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15, 207–218 (2004).
Engebrecht, J. Cell signaling in yeast sporulation. Biochem. Biophys. Res. Commun. 306, 325–328 (2003).
Kapranov, P., Routt, S.M., Bankaitis, V.A., de Bruijn, F.J. & Szczyglowski, K. Nodule-specific regulation of phosphatidylinositol transfer protein expression in Lotus japonicus. Plant Cell 13, 1369–1382 (2001).
Vincent, P. et al. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J. Cell Biol. 168, 801–812 (2005).
Anantharaman, V. & Aravind, L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 3 research0023 (2002).
Peterman, T.K., Ohol, Y.M., McReynolds, L.J. & Luna, E.J. Patellin1, a novel Sec14-like protein, localizes to the cell plate and binds phosphoinositides. Plant Physiol. 136, 3080–3094 discussion 3001–3002 (2004).
McLaughlin, S. & Murray, D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 (2005).
Yamniuk, A.P. & Vogel, H.J. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27, 33–57 (2004).
Weimar, W.R., Lane, P.W. & Sidman, R.L. Vibrator (vb): a spinocerebellar system degeneration with autosomal recessive inheritance in mice. Brain Res. 251, 357–364 (1982).
Hamilton, B.A. et al. The vibrator mutation causes neurodegeneration via reduced expression of PITP alpha: positional complementation cloning and extragenic suppression. Neuron 18, 711–722 (1997).
Alb, J.G., Jr. et al. Mice lacking phosphatidylinositol transfer protein-alpha exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J. Biol. Chem. 278, 33501–33518 (2003).
Xie, Y. et al. Phosphatidylinositol transfer protein-alpha in netrin-1-induced PLC signalling and neurite outgrowth. Nat. Cell Biol. 7, 1124–1132 (2005).
Giansanti, M.G. et al. The class I PITP giotto is required for Drosophila cytokinesis. Curr. Biol. 16, 195–201 (2006).
Gatt, M.K. & Glover, D.M. The Drosophila phosphatidylinositol transfer protein encoded by vibrator is essential to maintain cleavage-furrow ingression in cytokinesis. J. Cell Sci. 119, 2225–2235 (2006).
Milligan, S.C., Alb, J.G., Jr., Elagina, R.B., Bankaitis, V.A. & Hyde, D.R. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 139, 351–363 (1997).
Chang, J.T. et al. Mammalian homolog of Drosophila retinal degeneration B rescues the mutant fly phenotype. J. Neurosci. 17, 5881–5890 (1997).
Phillips, S.E., Ile, K.E., Boukhelifa, M., Huijbregts, R.P. & Bankaitis, V.A. Specific and nonspecific membrane-binding determinants cooperate in targeting phosphatidylinositol transfer protein beta-isoform to the mammalian trans-Golgi network. Mol. Biol. Cell 17, 2498–2512 (2006).
DeVries, K.J. et al. Fluorescently labeled phosphatidylinositol transfer protein isoforms (alpha and beta), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp. Cell Res. 227, 33–39 (1996).
Litvak, V., Dahan, N., Ramachandran, S., Sabanay, H. & Lev, S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat. Cell Biol. 7, 225–234 (2005).
Liljedahl, M. et al. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell 104, 409–420 (2001).
Baron, C.L. & Malhotra, V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 295, 325–328 (2002).
Jones, B. et al. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat. Genet. 34, 29–31 (2003).
Holthuis, J.C. & Levine, T.P. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6, 209–220 (2005).
Skinner, H.B., Alb, J.G., Jr., Whitters, E.A., Helmkamp, G.M., Jr. & Bankaitis, V.A. Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO J. 12, 4775–4784 (1993).
Nalbant, P., Hodgson, L., Kraynov, V., Toutchkine, A. & Hahn, K.M. Activation of endogenous Cdc42 visualized in living cells. Science 305, 1615–1619 (2004).
Raychaudhuri, S., Im, Y.J., Hurley, J.H. & Prinz, W.A. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 173, 107–119 (2006).
Yanagisawa, L.L. et al. Activity of specific lipid-regulated ADP ribosylation factor-GTPase-activating proteins is required for Sec14p-dependent Golgi secretory function in yeast. Mol. Biol. Cell 13, 2193–2206 (2002).
Lewis, S.M., Poon, P.P., Singer, R.A., Johnston, G.C. & Spang, A. The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol. Biol. Cell 15, 4064–4072 (2004).
Robinson, M. et al. The Gcs1 Arf-GAP mediates Snc1,2 v-SNARE retrieval to the Golgi in yeast. Mol. Biol. Cell 17, 1845–1858 (2006).
Lee, S.Y., Yang, J.S., Hong, W., Premont, R.T. & Hsu, V.W. ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J. Cell Biol. 168, 281–290 (2005).
Loewen, C.J., Roy, A. & Levine, T.P. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025–2035 (2003).
Acknowledgements
This work was supported by grants from the US National Institutes of Health to V.A.B. K.E.I. and G.S. were also supported by a Predoctoral Training Grant from the US National Institutes of Health and a Postdoctoral Fellowship from the Deutsche Forschungsgemeinschaft, respectively.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ile, K., Schaaf, G. & Bankaitis, V. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat Chem Biol 2, 576–583 (2006). https://doi.org/10.1038/nchembio835
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio835
This article is cited by
-
Chemical contrast for imaging living systems: molecular vibrations drive CARS microscopy
Nature Chemical Biology (2011)