Abstract
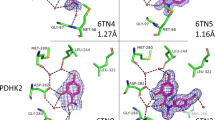
Fragment-based screens test multiple low–molecular weight molecules for binding to a target1,2,3,4. Fragments often bind with low affinities but typically have better ligand efficiencies (ΔGbind/heavy atom count) than traditional screening hits5. This efficiency, combined with accompanying atomic-resolution structures, has made fragments popular starting points for drug discovery programs2,6,7,8,9,10,11,12,13. Fragment-based design adopts a constructive strategy: affinity is enhanced either by cycles of functional-group addition or by joining two independent fragments together. The final inhibitor is expected to adopt the same geometry as the original fragment hit. Here we consider whether the inverse, deconstructive logic also applies—can one always parse a higher-affinity inhibitor into fragments that recapitulate the binding geometry of the larger molecule? Cocrystal structures of fragments deconstructed from a known β-lactamase inhibitor suggest that this is not always the case.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Erlanson, D.A., McDowell, R.S. & O'Brien, T. Fragment-based drug discovery. J. Med. Chem. 47, 3463–3482 (2004).
Erlanson, D.A., Wells, J.A. & Braisted, A.C. Tethering: fragment-based drug discovery. Annu. Rev. Biophys. Biomol. Struct. 33, 199–223 (2004).
Verdonk, M.L. & Hartshorn, M.J. Structure-guided fragment screening for lead discovery. Curr. Opin. Drug Discov. Devel. 7, 404–410 (2004).
Rees, D.C., Congreve, M., Murray, C.W. & Carr, R. Fragment-based lead discovery. Nat. Rev. Drug Discov. 3, 660–672 (2004).
Hopkins, A.L., Groom, C.R. & Alex, A. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today 9, 430–431 (2004).
Boehm, H.J. et al. Novel inhibitors of DNA gyrase: 3D structure based biased needle screening, hit validation by biophysical methods, and 3D guided optimization. A promising alternative to random screening. J. Med. Chem. 43, 2664–2674 (2000).
Shuker, S.B., Hajduk, P.J., Meadows, R.P. & Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531–1534 (1996).
Gill, A. New lead generation strategies for protein kinase inhibitors—fragment based screening approaches. Mini Rev. Med. Chem. 4, 301–311 (2004).
Card, G.L. et al. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design. Nat. Biotechnol. 23, 201–207 (2005).
Hartshorn, M.J. et al. Fragment-based lead discovery using X-ray crystallography. J. Med. Chem. 48, 403–413 (2005).
Huth, J.R., Sun, C., Sauer, D.R. & Hajduk, P.J. Utilization of NMR-derived fragment leads in drug design. Methods Enzymol. 394, 549–571 (2005).
Nienaber, V.L. et al. Discovering novel ligands for macromolecules using X-ray crystallographic screening. Nat. Biotechnol. 18, 1105–1108 (2000).
Blaney, J., Nienaber, V. & Burley, S.K. Fragment-based lead discovery and optimization using X-ray crystallography, computational chemistry, and high-throughput organic synthesis. in Fragment-based Approaches in Drug Discovery (eds. Jahnke, W. & Erlanson, D.) 215–248 (Wiley, New York, 2006).
Tondi, D., Morandi, F., Bonnet, R., Costi, M.P. & Shoichet, B.K. Structure-based optimization of a non-beta-lactam lead results in inhibitors that do not up-regulate beta-lactamase expression in cell culture. J. Am. Chem. Soc. 127, 4632–4639 (2005).
Usher, K.C., Shoichet, B.K., Blaszczak, L., Weston, G.S. & Remington, J.R. The three dimensional structure of AmpC β-lactamase from Escherichia coli bound to a transition-state analog: possible implications for the oxyanion hypothesis and for inhibitor design. Biochemistry 37, 16082–16092 (1998).
Powers, R.A. & Shoichet, B.K. Structure-based approach for binding site identification on AmpC beta-lactamase. J. Med. Chem. 45, 3222–3234 (2002).
Crichlow, G.V., Nukaga, M., Doppalapudi, V.R., Buynak, J.D. & Knox, J.R. Inhibition of class C beta-lactamases: structure of a reaction intermediate with a cephem sulfone. Biochemistry 40, 6233–6239 (2001).
Davis, A.M. & Teague, S.J. Hydrogen bonding, hydrophobic interactions, and failure of the rigid receptor hypothesis. Angew. Chem. Int. Edn Engl. 38, 736–749 (1999).
Stout, T.J., Sage, C.R. & Stroud, R.M. The additivity of substrate fragments in enzyme-ligand binding. Structure 6, 839–848 (1998).
Green, N.M. Avidin. Adv. Protein Chem. 29, 85–133 (1975).
Congreve, M.S. et al. Detection of ligands from a dynamic combinatorial library by X-ray crystallography. Angew. Chem. Int. Edn Engl. 42, 4479–4482 (2003).
Hajduk, P.J. et al. Discovery of potent nonpeptide inhibitors of stromelysin using SAR by NMR. J. Am. Chem. Soc. 119, 5818–5827 (1997).
Miller, B.G. & Wolfenden, R. Catalytic proficiency: the unusual case of OMP decarboxylase. Annu. Rev. Biochem. 71, 847–885 (2002).
Powers, R.A. et al. The complexed structure and antimicrobial activity of a non-beta-lactam inhibitor of AmpC beta-lactamase. Protein Sci. 8, 2330–2337 (1999).
Holton, J. & Alber, T. Automated protein crystal structure determination using ELVES. Proc. Natl. Acad. Sci. USA 101, 1537–1542 (2004).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
This work was supported by US National Institutes of Health grant GM59957 (to B.K.S.) and Ruth L. Kirschstein National Research Service Award fellowship GM076883 (to K.B). We thank B. Feng, J. Irwin, A. Graves and Y. Chen for reading the manuscript.
Author information
Authors and Affiliations
Contributions
K.B. and B.K.S. designed the experiments and wrote the manuscript together. K.B. did all of the actual experimental work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Electron density maps of fragments (PDF 440 kb)
Supplementary Table 1
Data collection and refinement statistics (PDF 30 kb)
Rights and permissions
About this article
Cite this article
Babaoglu, K., Shoichet, B. Deconstructing fragment-based inhibitor discovery. Nat Chem Biol 2, 720–723 (2006). https://doi.org/10.1038/nchembio831
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio831
This article is cited by
-
Nonadditivity in public and inhouse data: implications for drug design
Journal of Cheminformatics (2021)
-
Small molecule inhibitors against PD-1/PD-L1 immune checkpoints and current methodologies for their development: a review
Cancer Cell International (2021)
-
Design, synthesis and biological evaluation of non-covalent AmpC β-lactamases inhibitors
Medicinal Chemistry Research (2017)
-
Natural-product-derived fragments for fragment-based ligand discovery
Nature Chemistry (2013)
-
Computational fragment-based screening using RosettaLigand: the SAMPL3 challenge
Journal of Computer-Aided Molecular Design (2012)