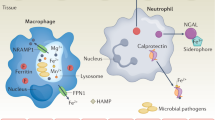
Abstract
Many bacteria, including numerous human pathogens, synthesize small molecules known as siderophores to scavenge iron. Enterobactin, a siderophore produced by enteric bacteria, is surprisingly ineffective as an iron-scavenging agent for bacteria growing in animals because of its hydrophobicity and its sequestration by the mammalian protein siderocalin, a component of the innate immune system. However, pathogenic strains of Escherichia coli and Salmonella use enzymes encoded by the iroA gene cluster to tailor enterobactin by glycosylation and linearization. The resulting modified forms of enterobactin, known as salmochelins, can evade siderocalin and are less hydrophobic than enterobactin, restoring this siderophore's iron-scavenging ability in mammals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Posey, J.E. & Gherardini, F.C. Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653 (2000).
Weinberg, E.D. The Lactobacillus anomaly: total iron abstinence. Perspect. Biol. Med. 40, 578–583 (1997).
Raymond, K.N., Dertz, E.A. & Kim, S.S. Enterobactin: an archetype for microbial iron transport. Proc. Natl Acad. Sci. USA 100, 3584–3588 (2003).
Andrews, S.C., Robinson, A.K. & Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 (2003).
Crosa, J.H. & Walsh, C.T. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66, 223–249 (2002).
Gehring, A.M., Mori, I. & Walsh, C.T. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37, 2648–2659 (1998).
Walsh, C., Liu, J., Rusnak, F. & Sakaitani, M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem. Rev. 90, 1105–1129 (1990).
Lambalot, R.H. et al. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem. Biol. 3, 923–936 (1996).
Furrer, J.L., Sanders, D.N., Hook-Barnard, I.G. & McIntosh, M.A. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44, 1225–1234 (2002).
Bleuel, C. et al. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 187, 6701–6707 (2005).
Crosa, J.H., Mey, A.R. & Payne, S.M. Iron Transport in Bacteria. (ASM Press, Washington, DC, 2004)
Annamalai, R., Jin, B., Cao, Z., Newton, S.M. & Klebba, P.E. Recognition of ferric catecholates by FepA. J. Bacteriol. 186, 3578–3589 (2004).
Buchanan, S.K. et al. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6, 56–63 (1999).
Faraldo-Gomez, J.D. & Sansom, M.S. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4, 105–116 (2003).
Lin, H., Fischbach, M.A., Liu, D.R. & Walsh, C.T. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127, 11075–11084 (2005).
Hantke, K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4, 172–177 (2001).
Harris, W.R. et al. Coordination chemistry of microbial iron transport compounds. 19. Stability constants and electrochemical behavior of ferric enterobactin and model complexes. J. Am. Chem. Soc. 101, 6097–6104 (1979).
Challis, G.L. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 6, 601–611 (2005).
Harris, W.R., Carrano, C.J. & Raymond, K.N. Coordination chemistry of microbial iron transport compounds. 16. Isolation, characterization, and formation constants of ferric aerobactin. J. Am. Chem. Soc. 110, 2722–2727 (1979).
Brock, J.H., Williams, P.H., Liceaga, J. & Wooldridge, K.G. Relative availability of transferrin-bound iron and cell-derived iron to aerobactin-producing and enterochelin-producing strains of Escherichia coli and to other microorganisms. Infect. Immun. 59, 3185–3190 (1991).
Der Vartanian, M. Differences in excretion and efficiency of the aerobactin and enterochelin siderophores in a bovine pathogenic strain of Escherichia coli. Infect. Immun. 56, 413–418 (1988).
Konopka, K., Bindereif, A. & Neilands, J.B. Aerobactin-mediated utilization of transferrin iron. Biochemistry 21, 6503–6508 (1982).
Williams, P.H. & Carbonetti, N.H. Iron, siderophores, and the pursuit of virulence: independence of the aerobactin and enterochelin iron uptake systems in Escherichia coli. Infect. Immun. 51, 942–947 (1986).
Goetz, D.H. et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10, 1033–1043 (2002).
Holmes, M.A., Paulsene, W., Jide, X., Ratledge, C. & Strong, R.K. Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure 13, 29–41 (2005).
Flo, T.H. et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 (2004).
Devireddy, L.R., Gazin, C., Zhu, X. & Green, M.R. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell 123, 1293–1305 (2005).
Devireddy, L.R., Teodoro, J.G., Richard, F.A. & Green, M.R. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science 293, 829–834 (2001).
Luo, M. et al. Enzymatic tailoring of the bacterial siderophore enterobactin alters membrane partitioning and iron acquisition. ACS Chem. Biol. (in the press).
Konopka, K. & Neilands, J.B. Effect of serum albumin on siderophore-mediated utilization of transferrin iron. Biochemistry 23, 2122–2127 (1984).
Bister, B. et al. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17, 471–481 (2004).
Koczura, R. & Kaznowski, A. Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog. 35, 197–202 (2003).
Baumler, A.J. et al. Identification of a new iron regulated locus of Salmonella typhi. Gene 183, 207–213 (1996).
Baumler, A.J. et al. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180, 1446–1453 (1998).
Foster, J.W. & Hall, H.K. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174, 4317–4323 (1992).
Patzer, S.I., Baquero, M.R., Bravo, D., Moreno, F. & Hantke, K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149, 2557–2570 (2003).
McClelland, M. et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856 (2001).
Welch, R.A. et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA 99, 17020–17024 (2002).
Grozdanov, L. et al. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186, 5432–5441 (2004).
Paulsen, I.T. et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074 (2003).
Dobrindt, U. et al. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70, 6365–6372 (2002).
Sorsa, L.J., Dufke, S., Heesemann, J. & Schubert, S. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 71, 3285–3293 (2003).
Snyder, J.A. et al. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72, 6373–6381 (2004).
Chen, Y.T. et al. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337, 189–198 (2004).
Lagos, R. et al. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42, 229–243 (2001).
Dean, C.R. & Poole, K. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol. Microbiol. 8, 1095–1103 (1993).
Hantke, K., Nicholson, G., Rabsch, W. & Winkelmann, G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl Acad. Sci. USA 100, 3677–3682 (2003).
Liu, J. & Mushegian, A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418–1431 (2003).
Walsh, C., Freel Meyers, C.L. & Losey, H.C. Antibiotic glycosyltransferases: antibiotic maturation and prospects for reprogramming. J. Med. Chem. 46, 3425–3436 (2003).
Fischbach, M.A., Lin, H., Liu, D.R. & Walsh, C.T. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl Acad. Sci. USA 102, 571–576 (2005).
Weymouth-Wilson, A.C. The role of carbohydrates in biologically active natural products. Nat. Prod. Rep. 14, 99–110 (1997).
Galm, U. et al. Antimicrobial and DNA gyrase-inhibitory activities of novel clorobiocin derivatives produced by mutasynthesis. Antimicrob. Agents Chemother. 48, 1307–1312 (2004).
Schlunzen, F. et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821 (2001).
Lafitte, D. et al. DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5′-methyl group of the noviose. Biochemistry 41, 7217–7223 (2002).
Janeway, C.A., Jr & Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002).
Quiros, L.M., Aguirrezabalaga, I., Olano, C., Mendez, C. & Salas, J.A. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 28, 1177–1185 (1998).
Walsh, C.T. Posttranslational Modification of Proteins: Expanding Nature's Inventory (Roberts and Co., Greenwood Village, Colorado, 2006).
Bililign, T., Griffith, B.R. & Thorson, J.S. Structure, activity, synthesis and biosynthesis of aryl-C-glycosides. Nat. Prod. Rep. 22, 742–760 (2005).
Hartmann, S. & Hofsteenge, J. Properdin, the positive regulator of complement, is highly C-mannosylated. J. Biol. Chem. 275, 28569–28574 (2000).
Hofsteenge, J., Blommers, M., Hess, D., Furmanek, A. & Miroshnichenko, O. The four terminal components of the complement system are C-mannosylated on multiple tryptophan residues. J. Biol. Chem. 274, 32786–32794 (1999).
Krieg, J. et al. C-Mannosylation of human RNase 2 is an intracellular process performed by a variety of cultured cells. J. Biol. Chem. 272, 26687–26692 (1997).
Perez-Vilar, J., Randell, S.H. & Boucher, R.C. C-Mannosylation of MUC5AC and MUC5B Cys subdomains. Glycobiology 14, 325–337 (2004).
Trefzer, A. et al. Function of glycosyltransferase genes involved in urdamycin A biosynthesis. Chem. Biol. 7, 133–142 (2000).
McMullen, M.D. et al. Quantitative trait loci and metabolic pathways. Proc. Natl Acad. Sci. USA 95, 1996–2000 (1998).
Zhu, M., Valdebenito, M., Winkelmann, G. & Hantke, K. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 151, 2363–2372 (2005).
Luo, M., Fadeev, E.A. & Groves, J.T. Membrane dynamics of the amphiphilic siderophore, acinetoferrin. J. Am. Chem. Soc. 127, 1726–1736 (2005).
Ratledge, C. & Ewing, M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology 142, 2207–2212 (1996).
Destoumieux-Garzon, D., Peduzzi, J. & Rebuffat, S. Focus on modified microcins: structural features and mechanisms of action. Biochimie 84, 511–519 (2002).
Li, Y.M., Milne, J.C., Madison, L.L., Kolter, R. & Walsh, C.T. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science 274, 1188–1193 (1996).
Bayro, M.J. et al. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J. Am. Chem. Soc. 125, 12382–12383 (2003).
Rosengren, K.J. et al. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 125, 12464–12474 (2003).
Semenova, E., Yuzenkova, Y., Peduzzi, J., Rebuffat, S. & Severinov, K. Structure-activity analysis of microcinJ25: distinct parts of the threaded lasso molecule are responsible for interaction with bacterial RNA polymerase. J. Bacteriol. 187, 3859–3863 (2005).
Wilson, K.A. et al. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J. Am. Chem. Soc. 125, 12475–12483 (2003).
Lagos, R., Wilkens, M., Vergara, C., Cecchi, X. & Monasterio, O. Microcin E492 forms ion channels in phospholipid bilayer membrane. FEBS Lett. 321, 145–148 (1993).
Thomas, X. et al. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279, 28233–28242 (2004).
Lagos, R., Villanueva, J.E. & Monasterio, O. Identification and properties of the genes encoding microcin E492 and its immunity protein. J. Bacteriol. 181, 212–217 (1999).
Strahsburger, E., Baeza, M., Monasterio, O. & Lagos, R. Cooperative uptake of microcin E492 by receptors FepA, Fiu, and Cir and inhibition by the siderophore enterochelin and its dimeric and trimeric hydrolysis products. Antimicrob. Agents Chemother. 49, 3083–3086 (2005).
Schubert, S., Fischer, D. & Heesemann, J. Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J. Bacteriol. 181, 6387–6395 (1999).
Trefzer, A. et al. Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic. Antimicrob. Agents Chemother. 46, 1174–1182 (2002).
Acknowledgements
Research in the authors' laboratories is supported by National Institutes of Health grants AI042738 (to C.T.W.) and GM065400 (to D.R.L.), and by the Howard Hughes Medical Institute (D.R.L.). M.A.F. is supported by a predoctoral fellowship from the Hertz Foundation, and H.L. is supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund. We are grateful to K. Hantke (Universität Tübingen) for helpful discussions.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Fischbach, M., Lin, H., Liu, D. et al. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol 2, 132–138 (2006). https://doi.org/10.1038/nchembio771
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio771
This article is cited by
-
Trait-Based Comparison of Coral and Sponge Microbiomes
Scientific Reports (2020)
-
You are what you eat: diet, health and the gut microbiota
Nature Reviews Gastroenterology & Hepatology (2019)
-
Comparative Genomics, Siderophore Production, and Iron Scavenging Potential of Root Zone Soil Bacteria Isolated from ‘Concord’ Grape Vineyards
Microbial Ecology (2019)
-
Multiple siderophores: bug or feature?
JBIC Journal of Biological Inorganic Chemistry (2018)
-
“Pumping iron”—how macrophages handle iron at the systemic, microenvironmental, and cellular levels
Pflügers Archiv - European Journal of Physiology (2017)