Abstract

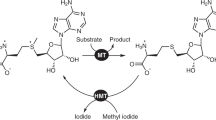
S-Adenosyl-L-methionine (AdoMet) is the major methyl donor for biological methylation reactions catalyzed by methyltransferases. We report the first chemical synthesis of AdoMet analogs with extended carbon chains replacing the methyl group and their evaluation as cofactors for all three classes of DNA methyltransferases. Extended groups containing a double or triple bond in the β position to the sulfonium center were transferred onto DNA in a catalytic and sequence-specific manner, demonstrating a high utility of such synthetic cofactors for targeted functionalization of biopolymers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Cantoni, G.L. J. Am. Chem. Soc. 74, 2942–2943 (1952).
Cheng, X. & Blumenthal, R.M. S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. 392 (World Scientific, Singapore, 1999).
Goll, M.G. & Bestor, T.H. Annu. Rev. Biochem. 74, 481–514 (2005).
Schlenk, F. & Dainko, J.L. Biochim. Biophys. Acta 385, 312–323 (1975).
Schlenk, F. in Biochemistry of Adenosylmethionine (eds. Salvatore, F., Borek, E., Zappia, V., Williams-Ashman, H.G. & Schlenk, F.) 3–17 (Columbia Univ. Press, New York, 1977).
De La Haba, G., Jamieson, G.A., Mudd, S.H. & Richards, H.H. J. Am. Chem. Soc. 81, 3975–3980 (1959).
Borchardt, R.T. & Wu, Y.S. J. Med. Chem. 19, 1099–1103 (1976).
Goedecke, K., Pignot, M., Goody, R.S., Scheidig, A.J. & Weinhold, E. Nat. Struct. Biol. 8, 121–125 (2001).
Klimasauskas, S., Kumar, S., Roberts, R.J. & Cheng, X. Cell 76, 357–369 (1994).
Merkiene, E., Vilkaitis, G. & Klimasauskas, S. Biol. Chem. 379, 569–571 (1998).
Ho, D.K., Wu, J.C., Santi, D.V. & Floss, H.G. Arch. Biochem. Biophys. 284, 264–269 (1991).
Schubert, H.L., Blumenthal, R.M. & Cheng, X. Trends Biochem. Sci. 28, 329–335 (2003).
Mi, S. & Roberts, R.J. Nucleic Acids Res. 21, 2459–2464 (1993).
Pljevaljcic, G., Schmidt, F. & Weinhold, E. Chem. Biochem. 5, 265–269 (2004).
Roberts, R.J., Vincze, T., Posfai, J. & Macelis, D. Nucleic Acids Res. 33, D230–D232 (2005).
Acknowledgements
The authors are grateful to E. Merkiene and M. Čaikovskij for constructing the Q82A variant of M.HhaI, F. Grygas and R. Gerasimaite for technical assistance and K. Glensk for preparing M.TaqI. We thank V. Gabelica and F. Rosu for performing high-resolution ESI-MS measurements at the Center for Analysis of Residues in Traces (CART), laboratory of E. De Pauw, University of Liège, Belgium. C.D. thanks the Deutsche Forschungsgemeinschaft for a stipend within the Graduiertenkolleg 440. This work was supported by grants from VolkswagenStiftung, the Howard Hughes Medical Institute and the Ministry of Science and Education of Lithuania.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
HPLC analysis of chemically and enzymatically synthesized S-adenosyl-L-ethionine. (PDF 363 kb)
Supplementary Fig. 2
Analysis of enzymatic transalkylation reactions by DNA MTases. (PDF 1450 kb)
Supplementary Fig. 3
Analysis of transalkylation products formed in duplex oligodeoxynucleotides. (PDF 472 kb)
Supplementary Fig. 4
Blockage of restriction endonuclease cleavage at overlapping sites on DNA by enzymatic incorporation of extended groups. (PDF 232 kb)
Supplementary Table 1
ESI-MS analysis of synthetic AdoMet analogs and modified nucleosides formed after enzymatic transalkylations with DNA MTases. (PDF 98 kb)
Rights and permissions
About this article
Cite this article
Dalhoff, C., Lukinavičius, G., Klimas̆auskas, S. et al. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat Chem Biol 2, 31–32 (2006). https://doi.org/10.1038/nchembio754
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio754
This article is cited by
-
Ribozyme for stabilized SAM analogue modifies RNA in cells
Nature Chemistry (2023)
-
A SAM analogue-utilizing ribozyme for site-specific RNA alkylation in living cells
Nature Chemistry (2023)
-
Biotechnological applications of S-adenosyl-methionine-dependent methyltransferases for natural products biosynthesis and diversification
Bioresources and Bioprocessing (2021)
-
Broadening the scope of biocatalytic C–C bond formation
Nature Reviews Chemistry (2020)
-
A metabolic labeling method detects m6A transcriptome-wide at single base resolution
Nature Chemical Biology (2020)