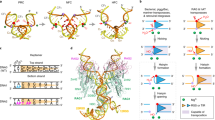
Abstract

The bacteriophage P1 Cre recombinase catalyzes site-specific recombination between 34-base-pair loxP sequences in a variety of topological contexts. This reaction is widely used to manipulate DNA molecules in applications ranging from benchtop cloning to genome modifications in transgenic animals. Despite the simple, highly symmetric nature of the Cre-loxP system, there is strong evidence that the reaction is asymmetric; the 'bottom' strands in the recombining loxP sites are preferentially exchanged before the 'top' strands. Here, we address the mechanistic basis for ordered strand exchange in the Cre-loxP recombination pathway. Using suicide substrates containing 5′-bridging phosphorothioate linkages at both cleavage sites, fluorescence resonance energy transfer between synapsed loxP sites and a Cre mutant that can cleave the bridging phosphorothioate linkage but not a normal phosphodiester linkage, we showed that preferential formation of a specific synaptic complex between loxP sites imposes ordered strand exchange during recombination and that synapsis stimulates cleavage of loxP sites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hoess, R.H., Ziese, M. & Sternberg, N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. USA 79, 3398–3402 (1982).
Sternberg, N., Hamilton, D., Austin, S., Yarmolinsky, M. & Hoess, R. Site-specific recombination and its role in the life cycle of bacteriophage P1. Cold Spring Harb. Symp. Quant. Biol. 45, 297–309 (1981).
Nash, H.A. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments. in Escherichia coli and Salmonella: Cellular and Molecular Biology (ed. Neidhardt, F.C. et al.) 2363–2376 (ASM Press, Washington, D.C., 1996).
Stark, W.M., Boocock, M.R. & Sherratt, D.J. Catalysis by site-specific recombinases. Trends Genet. 8, 432–439 (1992).
Azaro, M.A. & Landy, A. λ integrase and the λ Int family. in Mobile DNA II (eds. Craig, N.L., Craigie, R., Gellert, M. & Lambowitz, A.M.) 118–148 (ASM Press, Washington D.C., 2002).
Abremski, K. & Hoess, R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J. Biol. Chem. 259, 1509–1514 (1984).
Barre, F.X. et al. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 14, 2976–2988 (2000).
McCulloch, R., Coggins, L.W., Colloms, S.D. & Sherratt, D.J. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 13, 1844–1855 (1994).
Kitts, P.A. & Nash, H.A. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J. Mol. Biol. 204, 95–107 (1988).
Nunes-Düby, S., Matsumoto, L. & Landy, A. Site-specific recombination intermediates trapped with suicide substrates. Cell 50, 779–788 (1987).
Arciszewska, L.K. & Sherratt, D.J. Xer site-specific recombination in vitro. EMBO J. 14, 2112–2120 (1995).
Sauer, B. Chromosome manipulation by Cre-loxP recombination. in Mobile DNA II (eds. Craig, N.L., Craigie, R., Gellert, M. & Lambowitz, A.M.) 38–58 (ASM Press, Washington D.C., 2002).
Van Duyne, G.D. A structural view of Cre-loxP site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 30, 87–104 (2001).
Van Duyne, G.D. A structural view of tyrosine recombinase site-specific recombination. in Mobile DNA II (eds. Craig, N.L., Craigie, R., Gellert, M. & Lambowitz, A.M.) 93–117 (ASM Press, Washington DC, 2002).
Hoess, R., Wierzbicki, A. & Abremski, K. Isolation and characterization of intermediates in site-specific recombination. Proc. Natl. Acad. Sci. USA 84, 6840–6844 (1987).
Lee, L. & Sadowski, P.D. Sequence of the loxP site determines the order of strand exchange by the Cre recombinase. J. Mol. Biol. 326, 397–412 (2003).
Martin, S.S., Pulido, E., Chu, V.C., Lechner, T.S. & Baldwin, E.P. The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J. Mol. Biol. 319, 107–127 (2002).
Ennifar, E., Meyer, J.E., Buchholz, F., Stewart, A.F. & Suck, D. Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation. Nucleic Acids Res. 31, 5449–5460 (2003).
Lee, G. & Saito, I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216, 55–65 (1998).
Krogh, B.O. & Shuman, S. Catalytic mechanism of DNA topoisomerase IB. Mol. Cell 5, 1035–1041 (2000).
Burgin, A.B., Jr., Huizenga, B.N. & Nash, H.A. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 23, 2973–2979 (1995).
Burgin, A.B. & Nash, H.A. Suicide substrates reveal properties of the homology-dependent steps during integrative recombination of bacteriophage lambda. Curr. Biol. 5, 1312–1321 (1995).
Burgin, A.B. Jr. & Nash, H.A. Symmetry in the mechanism of bacteriophage lambda integrative recombination. Proc. Natl. Acad. Sci. USA 89, 9642–9646 (1992).
Xu, Y. & Kool, E.T. A novel 5′-iodonucleoside allows efficient nonenzymatic ligation of single-stranded and duplex DNAs. Tetrahedron Lett. 38, 5595–5598 (1997).
Miller, G.P. & Kool, E.T. A simple method for electrophilic functionalization of DNA. Org. Lett. 4, 3599–3601 (2002).
Ringrose, L. et al. Comparative kinetic analysis of FLP and Cre recombinases: mathematical models for DNA binding and recombination. J. Mol. Biol. 284, 363–384 (1998).
Abremski, K., Wierzbicki, A., Frommer, B. & Hoess, R.H. Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J. Biol. Chem. 261, 391–396 (1986).
Wierzbicki, A., Kendall, M., Abremski, K. & Hoess, R. A mutational analysis of the bacteriophage P1 recombinase Cre. J. Mol. Biol. 195, 785–794 (1987).
Cheng, C., Kussie, P., Pavletich, N. & Shuman, S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92, 841–850 (1998).
Guo, F., Gopaul, D.N. & Van Duyne, G.D. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl. Acad. Sci. USA 96, 7143–7148 (1999).
Guo, F., Gopaul, D.N. & Van Duyne, G.D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389, 40–46 (1997).
Gopaul, D.N., Guo, F. & Van Duyne, G.D. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 17, 4175–4187 (1998).
Ghosh, K., Lau, C.K., Guo, F., Segall, A.M. & Van Duyne, G.D. Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination. J. Biol. Chem. 280, 8290–8299 (2005).
Sherratt, D.J. & Wigley, D.B. Conserved themes but novel activities in recombinases and topoisomerases. Cell 93, 149–152 (1998).
Lee, L. & Sadowski, P.D. Directional resolution of synthetic holliday structures by the Cre recombinase. J. Biol. Chem. 276, 31092–31098 (2001).
Lee, L., Chu, L.C. & Sadowski, P.D. Cre induces an asymmetric DNA bend in its target loxP site. J. Biol. Chem. 278, 23118–23129 (2003).
Lee, L. & Sadowski, P.D. Strand selection by the tyrosine recombinases. in Progress in Nucleic Acid Research and Molecular Biology, Vol. 80 (ed. Moldave, K.) 1–42 (2005).
Voziyanov, Y., Lee, J., Whang, I. & Jayaram, M. Analyses of the first chemical step in Flp site-specific recombination: synapsis may not be a pre-requisite for strand cleavage. J. Mol. Biol. 256, 720–735 (1996).
Ghosh, K. & Van Duyne, G.D. Cre-loxP biochemistry. Methods 28, 374–383 (2002).
Gill, S.C. & von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326 (1989).
Clegg, R.M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211, 353–388 (1992).
Hoess, R.H., Wierzbicki, A. & Abremski, K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 14, 2287–2300 (1986).
Acknowledgements
We are grateful to R. Hoess for valuable discussions and insightful comments regarding this manuscript. This work was supported by a grant from the US National Institutes of Health. G.D.V. is Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
DNA Sequences. (PDF 486 kb)
Supplementary Fig. 2
Concentration of loxP (nM) (PDF 1866 kb)
Rights and permissions
About this article
Cite this article
Ghosh, K., Lau, CK., Gupta, K. et al. Preferential synapsis of loxP sites drives ordered strand exchange in Cre-loxP site-specific recombination. Nat Chem Biol 1, 275–282 (2005). https://doi.org/10.1038/nchembio733
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio733
This article is cited by
-
Site-specific bacterial chromosome engineering mediated by IntA integrase from Rhizobium etli
BMC Microbiology (2016)
-
Insights into the preferential order of strand exchange in the Cre/loxP recombinase system: impact of the DNA spacer flanking sequence and flexibility
Journal of Computer-Aided Molecular Design (2015)
-
Analysis of DNA processing reactions in bacterial conjugation by using suicide oligonucleotides
The EMBO Journal (2007)
-
Lox and Cre sandwich
Nature Chemical Biology (2005)