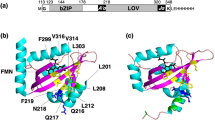
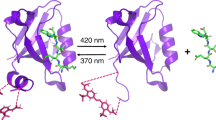
Abstract
Protein photosensors are versatile tools for studying ligand-regulated allostery and signaling. Fundamental to these processes is the amount of energy that can be provided by a photosensor to control downstream signaling events. Such regulation is exemplified by the phototropins—plant serine/threonine kinases that are activated by blue light via conserved LOV (light, oxygen and voltage) domains. The core photosensor of oat phototropin 1 is a LOV domain that interacts in a light-dependent fashion with an adjacent α-helix (Jα) to control kinase activity. We used solution NMR measurements to quantify the free energy of the LOV domain–Jα-helix binding equilibrium in the dark and lit states. These data indicate that light shifts this equilibrium by ∼3.8 kcal mol−1, thus quantifying the energy available through LOV-Jα for light-driven allosteric regulation. This study provides insight into the energetics of light sensing by phototropins and benchmark values for engineering photoswitchable systems based on the LOV-Jα interaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bhattacharyya, R.P., Remenyi, A., Yeh, B.J. & Lim, W.A. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75, 655–680 (2006).
Pawson, T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 (2004).
van der Horst, M.A. & Hellingwerf, K.J. Photoreceptor proteins, “Star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 37, 13–20 (2004).
Christie, J.M. Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58, 21–45 (2007).
Huala, E. et al. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278, 2120–2123 (1997).
Taylor, B.L. & Zhulin, I.B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63, 479–506 (1999).
Crosson, S., Rajagopal, S. & Moffat, K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42, 2–10 (2003).
Christie, J.M., Salomon, M., Nozue, K., Wada, M. & Briggs, W.R. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96, 8779–8783 (1999).
Crosson, S. & Moffat, K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14, 1067–1075 (2002).
Christie, J.M., Swartz, T.E., Bogomolni, R.A. & Briggs, W.R. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32, 205–219 (2002).
Fedorov, R. et al. Crystal structures and molecular mechanism of a light-induced signaling switch: The phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys. J. 84, 2474–2482 (2003).
Crosson, S. & Moffat, K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98, 2995–3000 (2001).
Harper, S.M., Neil, L.C. & Gardner, K.H. Structural basis of a phototropin light switch. Science 301, 1541–1544 (2003).
Zoltowski, B.D. et al. Conformational switching in the fungal light sensor vivid. Science 316, 1054–1057 (2007).
Halavaty, A.S. & Moffat, K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry 46, 14001–14009 (2007).
Möglich, A. & Moffat, K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol. 373, 112–126 (2007).
Corchnoy, S.B. et al. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem. 278, 724–731 (2003).
Harper, S.M., Christie, J.M. & Gardner, K.H. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry 43, 16184–16192 (2004).
Carver, J.P. & Richards, R.E. A general two-site solution for the chemical exchange produced dependence of T2 upon Carr-Purcell pulse separation. J. Magn. Reson. 6, 89–105 (1972).
Palmer, A.G. III, Kroenke, C.D. & Loria, J.P. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 339, 204–238 (2001).
Harper, S.M., Neil, L.C., Day, I.J., Hore, P.J. & Gardner, K.H. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. J. Am. Chem. Soc. 126, 3390–3391 (2004).
Genick, U.K., Soltis, S.M., Kuhn, P., Canestrelli, I.L. & Getzoff, E.D. Structure at 0.85 A resolution of an early protein photocycle intermediate. Nature 392, 206–209 (1998).
van der Horst, M.A., van Stokkum, I.H., Crielaard, W. & Hellingwerf, K.J. The role of the N-terminal domain of photoactive yellow protein in the transient partial unfolding during signalling state formation. FEBS Lett. 497, 26–30 (2001).
Harigai, M., Imamoto, Y., Kamikubo, H., Yamazaki, Y. & Kataoka, M. Role of an N-terminal loop in the secondary structural change of photoactive yellow protein. Biochemistry 42, 13893–13900 (2003).
Nolen, B., Taylor, S. & Ghosh, G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 (2004).
Buchwald, G. et al. Conformational switch and role of phosphorylation in PAK activation. Mol. Cell. Biol. 21, 5179–5189 (2001).
Gardino, A.K. & Kern, D. Functional dynamics of response regulators using NMR relaxation techniques. Methods Enzymol. 423, 149–165 (2007).
Volgraf, M. et al. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat. Chem. Biol. 2, 47–52 (2006).
Gorostiza, P. et al. Mechanisms of photoswitch conjugation and light activation of an ionotropic glutamate receptor. Proc. Natl. Acad. Sci. USA 104, 10865–10870 (2007).
Hausser, M. & Smith, S.L. Neuroscience: controlling neural circuits with light. Nature 446, 617–619 (2007).
Schroder-Lang, S. et al. Fast manipulation of cellular cAMP level by light in vivo. Nat. Methods 4, 39–42 (2007).
Dueber, J.E., Yeh, B.J., Chak, K. & Lim, W.A. Reprogramming control of an allosteric signaling switch through modular recombination. Science 301, 1904–1908 (2003).
Rosen, M.K. et al. Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol. 263, 627–636 (1996).
Lee, A.L., Urbauer, J.L. & Wand, A.J. Improved labeling strategy for 13C relaxation measurements of methyl groups in proteins. J. Biomol. NMR 9, 437–440 (1997).
Loria, J.P., Rance, M. & Palmer, A.G. III. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999).
Skrynnikov, N.R., Mulder, F.A., Hon, B., Dahlquist, F.W. & Kay, L.E. Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: application to methionine residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 123, 4556–4566 (2001).
Tollinger, M., Skrynnikov, N.R., Mulder, F.A., Forman-Kay, J.D. & Kay, L.E. Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 123, 11341–11352 (2001).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Johnson, B.A. & Blevins, R.A. NMRView - a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994).
Panchal, S.C., Bhavesh, N.S. & Hosur, R.V. Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: application to unfolded proteins. J. Biomol. NMR 20, 135–147 (2001).
Kay, L.E., Xu, G.Y. & Yamazaki, T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. A 109, 129–133 (1994).
Wittekind, M. & Mueller, L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. B 101, 201–205 (1993).
Grzesiek, S. & Bax, A. Correlating backbone amide and side-chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114, 6291–6293 (1992).
Rubinstenn, G. et al. Structural and dynamic changes of photoactive yellow protein during its photocycle in solution. Nat. Struct. Biol. 5, 568–570 (1998).
Cavanagh, J., Fairbrother, W.J., Palmer, A.G., Rance, M. & Skelton, N.J. Protein NMR Spectroscopy: Principles and Practice 1–687 (Academic Press, San Diego, 2007).
DeLano, W.L. The PyMOL Molecular Graphic System (DeLano Scientific, Palo Alto, California, USA, 2002).
Acknowledgements
Research was supported by grants from the Welch Foundation to M.K.R. (I–1544) and K.H.G. (I–1424) and by a grant from the Texas Higher Education Coordinating Board Advanced Technology Program (010019-0124-2003). We thank L.E. Kay (University of Toronto) for providing the pulse sequences and software for data acquisition and analyses, D.M. Korzhnev (University of Toronto) for help with experimental setup and data fitting and D. Kern (Brandeis University) for assistance with improved data acquisition methods. We also thank all members of the Rosen and Gardner labs for helpful discussions.
Author information
Authors and Affiliations
Contributions
X.Y., M.K.R. and K.H.G. designed experiments, analyzed data and wrote the paper, and X.Y. performed experiments.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 2980 kb)
Rights and permissions
About this article
Cite this article
Yao, X., Rosen, M. & Gardner, K. Estimation of the available free energy in a LOV2-Jα photoswitch. Nat Chem Biol 4, 491–497 (2008). https://doi.org/10.1038/nchembio.99
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.99
This article is cited by
-
Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine
Nature Communications (2022)
-
Development of light-responsive protein binding in the monobody non-immunoglobulin scaffold
Nature Communications (2020)
-
Photo-sensitive degron variants for tuning protein stability by light
BMC Systems Biology (2014)
-
Anisotropic energy flow and allosteric ligand binding in albumin
Nature Communications (2014)
-
Guiding lights: recent developments in optogenetic control of biochemical signals
Pflügers Archiv - European Journal of Physiology (2013)