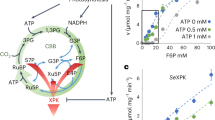
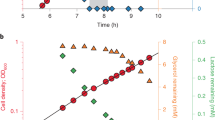
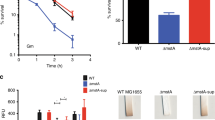
Abstract
Anapleurosis is the filling of the tricarboxylic acid cycle with four-carbon units. The common substrate for both anapleurosis and glucose phosphorylation in bacteria is the terminal glycolytic metabolite phosphoenolpyruvate (PEP). Here we show that Escherichia coli quickly and almost completely turns off PEP consumption upon glucose removal. The resulting buildup of PEP is used to quickly import glucose if it becomes available again. The switch-like termination of anapleurosis results from depletion of fructose-1,6-bisphosphate (FBP), an ultrasensitive allosteric activator of PEP carboxylase. E. coli expressing an FBP-insensitive point mutant of PEP carboxylase grow normally when glucose is steadily available. However, they fail to build up PEP upon glucose removal, grow poorly when glucose availability oscillates and suffer from futile cycling at the PEP node on gluconeogenic substrates. Thus, bacterial central carbon metabolism is intrinsically programmed with ultrasensitive allosteric regulation to enable rapid adaptation to changing environmental conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kotte, O., Zaugg, J.B. & Heinemann, M. Bacterial adaptation through distributed sensing of metabolic fluxes. Mol. Syst. Biol. 6, 355 (2010).
Christofk, H.R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008).
Sauer, U. & Eikmanns, B.J. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29, 765–794 (2005).
Feist, A.M. et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3, 121 (2007).
Siddiquee, K.A., Arauzo-Bravo, M.J. & Shimizu, K. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol. Lett. 235, 25–33 (2004).
Ishii, N. et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316, 593–597 (2007).
Fiehn, O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Brauer, M.J. et al. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA 103, 19302–19307 (2006).
Rabinowitz, J.D. & Kimball, E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79, 6167–6173 (2007).
Fischer, E. & Sauer, U. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J. Biol. Chem. 278, 46446–46451 (2003).
Salleh, H.M., Patel, M.A. & Woodard, R.W. Essential cysteines in 3-deoxy-D-manno-octulosonic acid 8-phosphate synthase from Escherichia coli: analysis by chemical modification and site-directed mutagenesis. Biochemistry 35, 8942–8947 (1996).
Kim, D.H. et al. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35, 4923–4928 (1996).
Al Zaid Siddiquee, K., Arauzo-Bravo, M.J. & Shimizu, K. Metabolic flux analysis of pykF gene knockout Escherichia coli based on C-13-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl. Microbiol. Biotechnol. 63, 407–417 (2004).
Haverkorn van Rijsewijk, B.R., Nanchen, A., Nallet, S., Kleijn, R.J. & Sauer, U. Large-scale 13C-flux analysis reveals distinct transcriptional control of respiratory and fermentative metabolism in Escherichia coli. Mol. Syst. Biol. 7, 477 (2011).
Peng, L. & Shimizu, K. Global metabolic regulation analysis for Escherichia coli K12 based on protein expression by 2-dimensional electrophoresis and enzyme activity measurement. Appl. Microbiol. Biotechnol. 61, 163–178 (2003).
Herrmann, K.M. The Shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 107, 7–12 (1995).
Sanwal, B.D. & Maeba, P. Regulation of activity of phosphoenolypyruvate carboxylase by fructose diphosphate. Biochem. Biophys. Res. Commun. 22, 194–199 (1966).
Izui, K., Matsumura, H., Furumoto, T. & Kai, Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu. Rev. Plant Biol. 55, 69–84 (2004).
Morikawa, M., Izui, K., Taguchi, M. & Katsuki, H. Regulation of Escherichia coli phosphoenolpyruvate carboxylase by multiple effectors in vivo. 1. Estimation of the activities in the cells grown on various compounds. J. Biochem. 87, 441–449 (1980).
Izui, K., Taguchi, M., Morikawa, M. & Katsuki, H. Regulation of Escherichia coli phosphoenolpyruvate carboxylase by multiple effectors in vivo. 2. Kinetic studies with a reaction system containing physiological concentrations of ligands. J. Biochem. 90, 1321–1331 (1981).
Yuan, J. & Rabinowitz, J.D. Differentiating metabolites formed from de novo synthesis versus macromolecule decomposition. J. Am. Chem. Soc. 129, 9294–9295 (2007).
Kodaki, T., Fujita, N., Kameshita, I., Izui, K. & Katsuki, H. Phosphoenolpyruvate carboxylase of Escherichia coli—specificity of some compounds as activators at the site for fructose-1,6-bisphosphate, one of the allosteric effectors. J. Biochem. 95, 637–642 (1984).
Garnak, M. & Reeves, H.C. Phosphorylation of isocitrate dehydrogenase of Escherichia coli. Science 203, 1111–1112 (1979).
Wohl, R.C. & Markus, G. Phosphoenolpyruvate carboxylase of Escherichia coli—purification and some properties. J. Biol. Chem. 247, 5785–5792 (1972).
Silverstein, R. & Willis, M.S. Concerted regulation in vitro of phosphoenolpyruvate carboxylase from Escherichia coli. J. Biol. Chem. 248, 8402–8407 (1973).
Takahashi-Terada, A. et al. Maize phosphoenolpyruvate carboxylase—mutations at the putative binding site for glucose 6-phosphate caused desensitization and abolished responsiveness to regulatory phosphorylation. J. Biol. Chem. 280, 11798–11806 (2005).
Oh, M.K., Rohlin, L., Kao, K.C. & Liao, J.C. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277, 13175–13183 (2002).
Bennett, B.D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009).
Zhao, J., Baba, T., Mori, H. & Shimizu, K. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab. Eng. 6, 164–174 (2004).
Schaub, J. & Reuss, M. In vivo dynamics of glycolysis in Escherichia coli shows need for growth-rate dependent metabolome analysis. Biotechnol. Prog. 24, 1402–1407 (2008).
Theobald, U., Mailinger, W., Baltes, M., Rizzi, M. & Reuss, M. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae. 1. Experimental observations. Biotechnol. Bioeng. 55, 305–316 (1997).
Zhu, T., Bailey, M.F., Angley, L.M., Cooper, T.F. & Dobson, R.C.J. The quaternary structure of pyruvate kinase type 1 from Escherichia coli at low nanomolar concentrations. Biochimie 92, 116–120 (2010).
Ogawa, T., Mori, H., Tomita, M. & Yoshino, M. Inhibitory effect of phosphoenolpyruvate on glycolytic enzymes in Escherichia coli. Res. Microbiol. 158, 159–163 (2007).
Kern, D. & Zuiderweg, E.R. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 13, 748–757 (2003).
Walsh, K. & Koshland, D.E. Branch point control by the phosphorylation state of isocitrate dehydrogenase—a quantitative examination of fluxes during a regulatory transition. J. Biol. Chem. 260, 8430–8437 (1985).
Stadtman, E.R. The story of glutamine synthetase regulation. J. Biol. Chem. 276, 44357–44364 (2001).
Kai, Y., Matsumura, H. & Izui, K. Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 414, 170–179 (2003).
Gonzalez, C.F. et al. Molecular basis of formaldehyde detoxification characterization of two S-formylglutathione hydrolases from Escherichia coli, Frmb and Yeig. J. Biol. Chem. 281, 14514–14522 (2006).
Brown, G. et al. Structural and biochemical characterization of the type II fructose-1,6-bisphosphatase GlpX from Escherichia coli. J. Biol. Chem. 284, 3784–3792 (2009).
Lee, M., Chan, C.W., Guss, J.M., Christopherson, R.I. & Maher, M.J. Dihydroorotase from Escherichia coli: loop movement and cooperativity between subunits. J. Mol. Biol. 348, 523–533 (2005).
Song, W.J. & Jackowski, S. Kinetics and regulation of pantothenate kinase from Escherichia coli. J. Biol. Chem. 269, 27051–27058 (1994).
Bhasin, M., Billinsky, J.L. & Palmer, D.R.J. Steady-state kinetics and molecular evolution of Escherichia coli MenD [(1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase], an anomalous thiamin diphosphate-dependent decarboxylase-carboligase. Biochemistry 42, 13496–13504 (2003).
Broglie, K.E. & Takahashi, M. Fluorescence studies of threonine-promoted conformational transitions in aspartokinase I using the substrate analogue 2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate. J. Biol. Chem. 258, 12940–12946 (1983).
Eisenstein, E., Yu, H.D. & Schwarz, F.P. Cooperative binding of the feedback modifiers isoleucine and valine to biosynthetic threonine deaminase from Escherichia coli. J. Biol. Chem. 269, 29423–29429 (1994).
Yuan, J., Bennett, B.D. & Rabinowitz, J.D. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat. Protoc. 3, 1328–1340 (2008).
Gutnick, D., Calvo, J.M., Klopotow, T. & Ames, B.N. Compounds which serve as sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100, 215–219 (1969).
Lu, W., Bennett, B.D. & Rabinowitz, J.D. Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871, 236–242 (2008).
Lu, W. et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 82, 3212–3221 (2010).
Kitagawa, M. et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete Set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299 (2005).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Acknowledgements
We thank Princeton University colleagues C. Doucette for the ppc deletion strain and A. Hottes for the pCA24N-ppc plasmid; J. Park for the flux calculations; A. Michaelis and Z. Gitai for the microscopy; D. Perlman, W. Lu, K. Saw and H. Shwe for protein mass spectrometry; and N. Wingreen for helpful discussions. This research was funded by US National Science Foundation (NSF) CAREER award MCB-0643859, Joint US Department of Energy–Air Force Office of Scientific Research award DOE DE-SC0002077–AFOSR FA9550-09-1-0580, the US National Institutes of Health Center for Quantitative Biology award P50 GM071508 and NSF grant CBET-0941143. M.L.R. was supported by NSF Graduate Research Fellowship DGE-0646086.
Author information
Authors and Affiliations
Contributions
Y.-F.X. and J.D.R. designed experiments, analyzed data and wrote the paper. D.A.-N. and M.L.R. performed preliminary experiments and contributed to discussion. X.-J.F. contributed to modeling.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 6895 kb)
Supplementary Data Set 1
Carbon starvation (XLSX 102 kb)
Rights and permissions
About this article
Cite this article
Xu, YF., Amador-Noguez, D., Reaves, M. et al. Ultrasensitive regulation of anapleurosis via allosteric activation of PEP carboxylase. Nat Chem Biol 8, 562–568 (2012). https://doi.org/10.1038/nchembio.941
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.941
This article is cited by
-
Dynamic fluctuations in a bacterial metabolic network
Nature Communications (2023)
-
A parallel glycolysis provides a selective advantage through rapid growth acceleration
Nature Chemical Biology (2023)
-
Near-equilibrium glycolysis supports metabolic homeostasis and energy yield
Nature Chemical Biology (2019)
-
The cyanobacterial ornithine–ammonia cycle involves an arginine dihydrolase
Nature Chemical Biology (2018)
-
Metabolite concentrations, fluxes and free energies imply efficient enzyme usage
Nature Chemical Biology (2016)