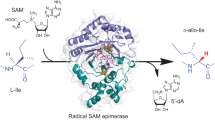
Abstract
Subtilosin A is a 35-residue, ribosomally synthesized bacteriocin encoded by the sbo-alb operon of Bacillus subtilis. It is composed of a head-to-tail circular peptide backbone that is additionally restrained by three unusual thioether bonds between three cysteines and the α-carbon of one threonine and two phenylalanines, respectively. In this study, we demonstrate that these bonds are synthesized by the radical S-adenosylmethionine enzyme AlbA, which is encoded by the sbo-alb operon and comprises two [4Fe-4S] clusters. One [4Fe-4S] cluster is coordinated by the prototypical CXXXCXXC motif and is responsible for the observed S-adenosylmethionine cleavage reaction, whereas the second [4Fe-4S] cluster is required for the generation of all three thioether linkages. On the basis of the obtained results, we propose a new radical mechanism for thioether bond formation. In addition, we show that AlbA-directed substrate transformation is leader-peptide dependent, suggesting that thioether bond formation is the first step during subtilosin A maturation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
16 April 2012
In the version of this article initially published, S-adenosyl methionine was drawn with a radical instead of a cation in Figure 4. The error has been corrected in the HTML and PDF versions of the article.
References
Hallen, H.E., Luo, H., Scott-Craig, J.S. & Walton, J.D. Gene family encoding the major toxins of lethal Amanita mushrooms. Proc. Natl. Acad. Sci. USA 104, 19097–19101 (2007).
Cascales, L. & Craik, D.J. Naturally occurring circular proteins: distribution, biosynthesis and evolution. Org. Biomol. Chem. 8, 5035–5047 (2010).
Maqueda, M. et al. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 32, 2–22 (2008).
Babasaki, K., Takao, T., Shimonishi, Y. & Kurahashi, K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. 98, 585–603 (1985).
Shelburne, C.E. et al. The spectrum of antimicrobial activity of the bacteriocin subtilosin A. J. Antimicrob. Chemother. 59, 297–300 (2007).
Sutyak, K.E. et al. Spermicidal activity of the safe natural antimicrobial peptide subtilosin. Infect. Dis. Obstet. Gynecol. 2008, 540758 (2008).
Silkin, L., Hamza, S., Kaufman, S., Cobb, S.L. & Vederas, J.C. Spermicidal bacteriocins: lacticin 3147 and subtilosin A. Bioorg. Med. Chem. Lett. 18, 3103–3106 (2008).
Huang, T. et al. Isolation of a variant of subtilosin A with hemolytic activity. J. Bacteriol. 191, 5690–5696 (2009).
Kawulka, K. et al. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to α-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 125, 4726–4727 (2003).
Kawulka, K.E. et al. Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to α-carbon cross-links: formation and reduction of α-thio-α-amino acid derivatives. Biochemistry 43, 3385–3395 (2004).
Liu, W.-T. et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 107, 16286–16290 (2010).
Lee, H., Churey, J.J. & Worobo, R.W. Biosynthesis and transcriptional analysis of thurincin H, a tandem repeated bacteriocin genetic locus, produced by Bacillus thuringiensis SF361. FEMS Microbiol. Lett. 299, 205–213 (2009).
Sit, C.S., van Belkum, M.J., McKay, R.T., Worobo, R.W. & Vederas, J.C. The 3D solution structure of thurincin H, a cacteriocin with four sulfur to α-carbon crosslinks. Angew. Chem. Int. Edn Engl. 50, 8718–8721 (2011).
Rea, M.C. et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. USA 107, 9352–9357 (2010).
Sit, C.S., McKay, R.T., Hill, C., Ross, R.P. & Vederas, J.C. The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to α-carbon cross-links. J. Am. Chem. Soc. 133, 7680–7683 (2011).
Zheng, G., Yan, L.Z., Vederas, J.C. & Zuber, P. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 181, 7346–7355 (1999).
Zheng, G., Hehn, R. & Zuber, P. Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 182, 3266–3273 (2000).
Nakano, M.M., Zheng, G. & Zuber, P. Dual control of sbo-alb operon expression by the Spo0 and ResDE systems of signal transduction under anaerobic conditions in Bacillus subtilis. J. Bacteriol. 182, 3274–3277 (2000).
Albano, M. et al. The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J. Bacteriol. 187, 2010–2019 (2005).
González-Pastor, J.E., Hobbs, E.C. & Losick, R. Cannibalism by sporulating bacteria. Science 301, 510–513 (2003).
Sofia, H.J., Chen, G., Hetzler, B., Reyes-Spindola, J. & Miller, N. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 (2001).
Frey, P.A., Hegeman, A.D. & Ruzicka, F.J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 (2008).
Duschene, K.S., Veneziano, S.E., Silver, S.C. & Broderick, J.B. Control of radical chemistry in the AdoMet radical enzymes. Curr. Opin. Chem. Biol. 13, 74–83 (2009).
Chirpich, T.P., Zappia, V., Costilow, R.N. & Barker, H.A. Lysine 2,3-aminomutase. Purification and properties of a pyridoxal phosphate and S-adenosylmethionine-activated enzyme. J. Biol. Chem. 245, 1778–1789 (1970).
Layer, G. et al. Structural and functional comparison of HemN to other radical SAM enzymes. Biol. Chem. 386, 971–980 (2005).
Guianvarc'h, D., Florentin, D., Tse Sum Bui, B., Nunzi, F. & Marquet, A. Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical. Biochem. Biophys. Res. Commun. 236, 402–406 (1997).
Ugulava, N.B., Gibney, B.R. & Jarrett, J.T. Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry 40, 8343–8351 (2001).
Berkovitch, F. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 (2004).
Miller, J.R. et al. Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry 39, 15166–15178 (2000).
Cicchillo, R.M. et al. Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry 43, 6378–6386 (2004).
Santamaria-Araujo, J.A. et al. The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J. Biol. Chem. 279, 15994–15999 (2004).
Hänzelmann, P. & Schindelin, H. Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc. Natl. Acad. Sci. USA 103, 6829–6834 (2006).
Pierrel, F., Douki, T., Fontecave, M. & Atta, M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J. Biol. Chem. 279, 47555–47563 (2004).
Hernández, H.L. et al. MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry 46, 5140–5147 (2007).
Lee, K.-H. et al. Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry 48, 10162–10174 (2009).
Yokoyama, K., Numakura, M., Kudo, F., Ohmori, D. & Eguchi, T. Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. J. Am. Chem. Soc. 129, 15147–15155 (2007).
Grove, T.L., Ahlum, J.H., Sharma, P., Krebs, C. & Booker, S.J. A consensus mechanism for radical SAM-dependent dehydrogenation? BtrN contains two [4Fe-4S] clusters. Biochemistry 49, 3783–3785 (2010).
Fang, Q., Peng, J. & Dierks, T. Post-translational formylglycine modification of bacterial sulfatases by the radical S-adenosylmethionine protein AtsB. J. Biol. Chem. 279, 14570–14578 (2004).
Grove, T.L., Lee, K.-H., St. Clair, J., Krebs, C. & Booker, S.J. In vitro characterization of AtsB, a radical SAM formylglycine-generating enzyme that contains three [4Fe-4S] clusters. Biochemistry 47, 7523–7538 (2008).
Hagen, K. & Watson, A. Synthetic routes to iron sulfide (Fe2S2, Fe3S4, Fe4S4, and Fe6S9), clusters from the common precursor tetrakis(ethanethiolate)ferrate2– ion ([Fe(SC2H5)4]2–): structures and properties of [Fe3S4(SR)4]3– and bis(ethanethiolate)nonathioxohexaferrate4– ion ([Fe6S9(SC2H5)2]4–), examples of the newest types of Fe-S-SR clusters. J. Am. Chem. Soc. 105, 3905–3913 (1983).
Külzer, R., Pils, T., Kappl, R., Hüttermann, J. & Knappe, J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J. Biol. Chem. 273, 4897–4903 (1998).
Ugulava, N.B., Gibney, B.R. & Jarrett, J.T. Iron-sulfur cluster interconversions in biotin synthase: dissociation and reassociation of iron during conversion of [2Fe-2S] to [4Fe-4S] clusters. Biochemistry 39, 5206–5214 (2000).
Duschene, K.S. & Broderick, J.B. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 584, 1263–1267 (2010).
Chatterjee, A. et al. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol. 4, 758–765 (2008).
Murphy, K. et al. Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PLoS ONE 6, e20852 (2011).
Oman, T.J. & van der Donk, W.A. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 6, 9–18 (2010).
Xie, L., Miller, L., Chatterjee, C. & Averin, O. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science 303, 679–681 (2004).
Chatterjee, C., Paul, M., Xie, L. & van der Donk, W.A. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–684 (2005).
Roach, P.L., Clifton, I., Hensgens, C. & Shibata, N. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 387, 827–830 (1997).
Layer, G., Verfürth, K., Mahlitz, E. & Jahn, D. Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli. J. Biol. Chem. 277, 34136–34142 (2002).
Acknowledgements
We would like to thank N. Fritzler and J. Bamberger for practical realization of the HPLC-HRMS measurements. In addition, we would like to thank the whole Marahiel group for fruitful discussions. Furthermore, we gratefully acknowledge financial support from the Deutsche Forschungsgemeinschaft and from the LOEWE Center for Synthetic Microbiology.
Author information
Authors and Affiliations
Contributions
T.A.K. and M.A.M. initiated the project. M.J.G., L.F. and T.A.K. conceived the experiments. M.J.G. performed preliminary experiments for the characterization of AlbA. L.F. performed most experiments. M.J.G. and L.F. prepared the figures. O.B. carried out the EPR measurements. U.L. designed the HPLC-MS gradients and carried out the measurements. A.S. cloned, expressed and purified AlbA. L.F. and T.A.K. analyzed and interpreted the obtained data. L.F., T.A.K. and M.A.M. coordinated the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 11619 kb)
Rights and permissions
About this article
Cite this article
Flühe, L., Knappe, T., Gattner, M. et al. The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat Chem Biol 8, 350–357 (2012). https://doi.org/10.1038/nchembio.798
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.798
This article is cited by
-
Bicyclostreptins are radical SAM enzyme-modified peptides with unique cyclization motifs
Nature Chemical Biology (2022)
-
Maturation strategy influences expression levels and cofactor occupancy in Fe–S proteins
JBIC Journal of Biological Inorganic Chemistry (2022)
-
Characterizing SPASM/twitch Domain-Containing Radical SAM Enzymes by EPR Spectroscopy
Applied Magnetic Resonance (2022)
-
Culture and genome-based analysis of four soil Clostridium isolates reveal their potential for antimicrobial production
BMC Genomics (2021)
-
Biomanufacturing process for the production of bacteriocins from Bacillaceae family
Bioresources and Bioprocessing (2020)