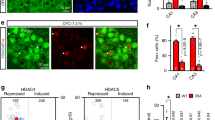
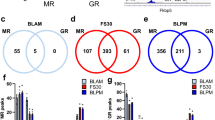
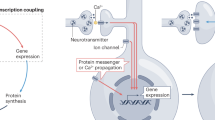
Abstract
The transcription factor cyclic AMP–response element binding protein (CREB) is a key regulator of many neuronal processes, including brain development, circadian rhythm and long-term memory. Studies of CREB have focused on its phosphorylation, although the diversity of CREB functions in the brain suggests additional forms of regulation. Here we expand on a chemoenzymatic strategy for quantifying glycosylation stoichiometries to characterize the functional roles of CREB glycosylation in neurons. We show that CREB is dynamically modified with an O-linked β-N-acetyl-D-glucosamine sugar in response to neuronal activity and that glycosylation represses CREB-dependent transcription by impairing its association with CREB-regulated transcription coactivator (CRTC; also known as transducer of regulated CREB activity). Blocking glycosylation of CREB alters cellular function and behavioral plasticity, enhancing both axonal and dendritic growth and long-term memory consolidation. Our findings demonstrate a new role for O-glycosylation in memory formation and provide a mechanistic understanding of how glycosylation contributes to critical neuronal functions. Moreover, we identify a previously unknown mechanism for the regulation of activity-dependent gene expression, neural development and memory.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lonze, B.E., Riccio, A., Cohen, S. & Ginty, D.D. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron 34, 371–385 (2002).
Carlezon, W.A. Jr. et al. Regulation of cocaine reward by CREB. Science 282, 2272–2275 (1998).
Kida, S. et al. CREB required for the stability of new and reactivated fear memories. Nat. Neurosci. 5, 348–355 (2002).
Kornhauser, J.M. et al. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron 34, 221–233 (2002).
Gau, D. et al. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron 34, 245–253 (2002).
Lonze, B.E. & Ginty, D.D. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623 (2002).
Shaywitz, A.J. & Greenberg, M.E. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68, 821–861 (1999).
Barco, A., Jancic, D. & Kandel, E.R. CREB-dependent transcription and synaptic plasticity. in Transcriptional Regulation by Neuronal Activity (ed. Dudek, S.M.) 127–154 (Springer US, 2008).
Chrivia, J.C. et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859 (1993).
Deisseroth, K. & Tsien, R.W. Dynamic multiphosphorylation passwords for activity-dependent gene expression. Neuron 34, 179–182 (2002).
Conkright, M.D. et al. TORCs: transducers of regulated CREB activity. Mol. Cell 12, 413–423 (2003).
Hart, G.W., Housley, M.P. & Slawson, C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 (2007).
Rexach, J.E., Clark, P.M. & Hsieh-Wilson, L.C. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat. Chem. Biol. 4, 97–106 (2008).
Love, D.C. & Hanover, J.A. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci. STKE 2005, re13 (2005).
Khidekel, N., Ficarro, S.B., Peters, E.C. & Hsieh-Wilson, L.C. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. USA 101, 13132–13137 (2004).
Vosseller, K. et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 (2006).
Greengard, P. The neurobiology of slow synaptic transmission. Science 294, 1024–1030 (2001).
Tallent, M.K. et al. In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J. Biol. Chem. 284, 174–181 (2009).
Rengifo, J., Gibson, C.J., Winkler, E., Collin, T. & Ehrlich, B.E. Regulation of the inositol 1,4,5-trisphosphate receptor type I by O-GlcNAc glycosylation. J. Neurosci. 27, 13813–13821 (2007).
Francisco, H. et al. O-GlcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev. Neurobiol. 69, 162–173 (2009).
Liu, F., Iqbal, K., Grundke-Iqbal, I., Hart, G.W. & Gong, C.X. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc. Natl. Acad. Sci. USA 101, 10804–10809 (2004).
Ohtsubo, K. & Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 (2006).
Shafi, R. et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97, 5735–5739 (2000).
Rexach, J.E. et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 6, 645–651 (2010).
Lamarre-Vincent, N. & Hsieh-Wilson, L.C. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J. Am. Chem. Soc. 125, 6612–6613 (2003).
Hagiwara, M. et al. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol. Cell. Biol. 13, 4852–4859 (1993).
Zhou, Z. et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52, 255–269 (2006).
Wu, G.Y., Deisseroth, K. & Tsien, R.W. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. USA 98, 2808–2813 (2001).
Plath, N. et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52, 437–444 (2006).
Egan, M.F. et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 112, 257–269 (2003).
Wayman, G.A. et al. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50, 897–909 (2006).
Ohshima, T. et al. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc. Natl. Acad. Sci. USA 93, 11173–11178 (1996).
Fleischmann, A. et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J. Neurosci. 23, 9116–9122 (2003).
Aguado, F. et al. The CREB/CREM transcription factors negatively regulate early synaptogenesis and spontaneous network activity. J. Neurosci. 29, 328–333 (2009).
Tucker, K.L., Meyer, M. & Barde, Y.A. Neurotrophins are required for nerve growth during development. Nat. Neurosci. 4, 29–37 (2001).
Zhang, L.I. & Poo, M.M. Electrical activity and development of neural circuits. Nat. Neurosci. 4 Suppl: 1207–1214 (2001).
Viosca, J., Lopez de Armentia, M., Jancic, D. & Barco, A. Enhanced CREB-dependent gene expression increases the excitability of neurons in the basal amygdala and primes the consolidation of contextual and cued fear memory. Learn. Mem. 16, 193–197 (2009).
Bourtchuladze, R. et al. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68 (1994).
Han, J.H. et al. Neuronal competition and selection during memory formation. Science 316, 457–460 (2007).
Bartsch, D., Casadio, A., Karl, K.A., Serodio, P. & Kandel, E.R. CREB1 encodes a nuclear activator, a repressor and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell 95, 211–223 (1998).
Zhou, Y. et al. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat. Neurosci. 12, 1438–1443 (2009).
Bourtchouladze, R. et al. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn. Mem. 5, 365–374 (1998).
Xiong, W. et al. Anisomycin activates p38 MAP kinase to induce LTD in mouse primary visual cortex. Brain Res. 1085, 68–76 (2006).
Yang, X. et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 (2008).
Dentin, R., Hedrick, S., Xie, J., Yates, J. III & Montminy, M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science 319, 1402–1405 (2008).
Comerford, K.M. et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. USA 100, 986–991 (2003).
Lu, Q., Hutchins, A.E., Doyle, C.M., Lundblad, J.R. & Kwok, R.P. Acetylation of cAMP-responsive element-binding protein (CREB) by CREB-binding protein enhances CREB-dependent transcription. J. Biol. Chem. 278, 15727–15734 (2003).
Yang, X., Zhang, F. & Kudlow, J.E. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 (2002).
Gambetta, M.C., Oktaba, K. & Muller, J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 (2009).
Wearne, S.L. et al. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience 136, 661–680 (2005).
Acknowledgements
We thank M. Antoniou (King's College London School of Medicine) for the pA2UCOE-EGFP construct, M. Greenberg (Harvard University) for the pLEMPRA-GOI and pLLX-shRNA constructs, G. Hart (The John Hopkins University School of Medicine) for the OGT-specific antibody, S. Josselyn (University of Toronto) for the p1005-CREB construct, R. Lansford (California Institute of Technology) for the pLenti PGK-H2B-mCherry construct, R. Malenka (Stanford University) and X. Yu (Shanghai Institutes for Biological Sciences) for the pcDNA3-Dkk-1-Flag and Ncad(intra) constructs, P. Qasba (US National Cancer Institute) for the Y289L GalT construct and L. Wells (University of Georgia) for the pDEST-HA-OGA construct. We thank S.-H. Yu (California Institute of Technology) for synthesizing the UDP-ketogalactose substrate and D. Anderson (California Institute of Technology) for providing the fear conditioning apparatus. We thank A. Silva for a critical reading of the manuscript. This work was supported by grants from the US National Institutes of Health (R01 GM084724 to L.C.H.-W., F31 NS056525 to J.E.R. and National Research Service Award Training Grant 5T32 GM07737 to P.M.C.).
Author information
Authors and Affiliations
Contributions
L.C.H.-W. designed, directed and coordinated the project. P.M.C. and J.E.R. designed and performed the experiments except where otherwise noted. D.E.M. and E.C.P. performed the MS analyses; R.L.N. prepared the HSV. P.M.C., J.E.R. and L.C.H.-W. wrote the manuscript, and all authors participated in editing it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 8224 kb)
Rights and permissions
About this article
Cite this article
Rexach, J., Clark, P., Mason, D. et al. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol 8, 253–261 (2012). https://doi.org/10.1038/nchembio.770
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.770
This article is cited by
-
Modulation of synaptic transmission through O-GlcNAcylation
Molecular Brain (2024)
-
Interoceptive regulation of skeletal tissue homeostasis and repair
Bone Research (2023)
-
O-GlcNAcylation determines the translational regulation and phase separation of YTHDF proteins
Nature Cell Biology (2023)
-
Humulus lupulus L. extract and its active constituent xanthohumol attenuate oxidative stress and nerve injury induced by iron overload via activating AKT/GSK3β and Nrf2/NQO1 pathways
Journal of Natural Medicines (2023)
-
Loss of O-GlcNAcylation on MeCP2 at Threonine 203 Leads to Neurodevelopmental Disorders
Neuroscience Bulletin (2022)