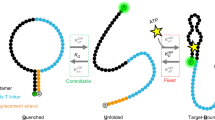
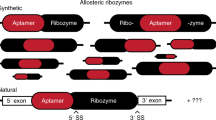
Abstract
Aptamers are useful for allosteric regulation because they are nucleic acid–based structures in which ligand binding induces conformational changes that may alter the function of a connected oligonucleotide at a distant site. Through this approach, a specific input is efficiently converted into an altered output. This property makes these biomolecules ideally suited to function as sensors or switches in biochemical assays or inside living cells. The ability to select oligonucleotide-based recognition elements in vitro in combination with the availability of nucleic acids with enzymatic activity has led to the development of a wide range of engineered allosteric aptasensors and aptazymes. Here, we discuss recent progress in the screening, design and diversity of these conformational switching oligonucleotides. We cover their application in vitro and for regulating gene expression in both prokaryotes and eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Golynskiy, M.V., Koay, M.S., Vinkenborg, J.L. & Merkx, M. Engineering protein switches: sensors, regulators, and spare parts for biology and biotechnology. ChemBioChem 12, 353–361 (2011).
Mayer, G. The chemical biology of aptamers. Angew. Chem. Int. Edn Engl. 48, 2672–2689 (2009).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Ellington, A.D. & Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Famulok, M., Hartig, J.S. & Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 107, 3715–3743 (2007).
Famulok, M. & Mayer, G. Aptamers as tools in molecular biology and immunology. Curr. Top. Microbiol. Immunol. 243, 123–136 (1999).
Tang, J. & Breaker, R.R. Rational design of allosteric ribozymes. Chem. Biol. 4, 453–459 (1997).
Stojanovic, M.N. & Kolpashchikov, D.M. Modular aptameric sensors. J. Am. Chem. Soc. 126, 9266–9270 (2004).
Nudler, E. Flipping riboswitches. Cell 126, 19–22 (2006).
Winkler, W.C. & Breaker, R.R. Genetic control by metabolite-binding riboswitches. ChemBioChem 4, 1024–1032 (2003).
Liu, J., Cao, Z. & Lu, Y. Functional nucleic acid sensors. Chem. Rev. 109, 1948–1998 (2009).
Tombelli, S., Minunni, M. & Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 20, 2424–2434 (2005).
Soukup, G.A. & Breaker, R.R. Nucleic acid molecular switches. Trends Biotechnol. 17, 469–476 (1999).
Famulok, M. Allosteric aptamers and aptazymes as probes for screening approaches. Curr. Opin. Mol. Ther. 7, 137–143 (2005).
Mayer, G., Raddatz, M.S., Grunwald, J.D. & Famulok, M. RNA ligands that distinguish metabolite-induced conformations in the TPP riboswitch. Angew. Chem. Int. Edn Engl. 46, 557–560 (2007).
Rentmeister, A., Mayer, G., Kuhn, N. & Famulok, M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 35, 3713–3722 (2007).
Vaish, N.K. et al. Monitoring post-translational modification of proteins with allosteric ribozymes. Nat. Biotechnol. 20, 810–815 (2002).
Srinivasan, J. et al. ADP-specific sensors enable universal assay of protein kinase activity. Chem. Biol. 11, 499–508 (2004).
Chiuman, W. & Li, Y. Simple fluorescent sensors engineered with catalytic DNA 'MgZ' based on a non-classic allosteric design. PLoS ONE 2, e1224 (2007).
Sekella, P.T., Rueda, D. & Walter, N.G. A biosensor for theophylline based on fluorescence detection of ligand-induced hammerhead ribozyme cleavage. RNA 8, 1242–1252 (2002).
Hartig, J.S. et al. Protein-dependent ribozymes report molecular interactions in real time. Nat. Biotechnol. 20, 717–722 (2002).
Najafi-Shoushtari, S.H. & Famulok, M. DNA aptamer-mediated regulation of the hairpin ribozyme by human alpha-thrombin. Blood Cells Mol. Dis. 38, 19–24 (2007).
Najafi-Shoushtari, S.H., Mayer, G. & Famulok, M. Sensing complex regulatory networks by conformationally controlled hairpin ribozymes. Nucleic Acids Res. 32, 3212–3219 (2004).
Yin, P., Choi, H.M., Calvert, C.R. & Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 451, 318–322 (2008).
Choi, H.M. et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 28, 1208–1212 (2010).
Liu, J. & Lu, Y. Adenosine-dependent assembly of aptazyme-functionalized gold nanoparticles and its application as a colorimetric biosensor. Anal. Chem. 76, 1627–1632 (2004).
Liu, J. & Lu, Y. Smart nanomaterials responsive to multiple chemical stimuli with controllable cooperativity. Adv. Mater. 18, 1667–1671 (2006).
Liu, J., Mazumdar, D. & Lu, Y. A simple and sensitive 'dipstick' test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew. Chem. Int. Edn Engl. 45, 7955–7959 (2006). This application involves gold nanoparticles functionalized with allosteric aptamers in a 'dipstick' test to detect cocaine levels in serum with the naked eye.
Mazumdar, D., Liu, J., Lu, G., Zhou, J. & Lu, Y. Easy-to-use dipstick tests for detection of lead in paints using non-cross-linked gold nanoparticle-DNAzyme conjugates. Chem. Commun. (Camb.) 46, 1416–1418 (2010).
Helm, M., Petermeier, M., Ge, B., Fiammengo, R. & Jaschke, A. Allosterically activated Diels-Alder catalysis by a ribozyme. J. Am. Chem. Soc. 127, 10492–10493 (2005).
Robertson, M.P. & Ellington, A.D. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 17, 62–66 (1999).
Cho, E.J., Yang, L., Levy, M. & Ellington, A.D. Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. J. Am. Chem. Soc. 127, 2022–2023 (2005).
Kim, D.E. & Joyce, G.F. Cross-catalytic replication of an RNA ligase ribozyme. Chem. Biol. 11, 1505–1512 (2004).
Lincoln, T.A. & Joyce, G.F. Self-sustained replication of an RNA enzyme. Science 323, 1229–1232 (2009).
Lam, B.J. & Joyce, G.F. Autocatalytic aptazymes enable ligand-dependent exponential amplification of RNA. Nat. Biotechnol. 27, 288–292 (2009). Elegant example in which loop replacement yields allosteric regulation of a self-replicating RNA, providing an accumulative output in response to ligand without requiring a polymerase.
Lam, B.J. & Joyce, G.F. An isothermal system that couples ligand-dependent catalysis to ligand-independent exponential amplification. J. Am. Chem. Soc. 133, 3191–3197 (2011).
Travascio, P., Li, Y. & Sen, D. DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem. Biol. 5, 505–517 (1998).
Pelossof, G., Tel-Vered, R., Elbaz, J. & Willner, I. Amplified biosensing using the horseradish peroxidase-mimicking DNAzyme as an electrocatalyst. Anal. Chem. 82, 4396–4402 (2010).
Teller, C., Shimron, S. & Willner, I. Aptamer-DNAzyme hairpins for amplified biosensing. Anal. Chem. 81, 9114–9119 (2009).
Li, T., Wang, E. & Dong, S. Lead(II)-induced allosteric G-quadruplex DNAzyme as a colorimetric and chemiluminescence sensor for highly sensitive and selective Pb2+ detection. Anal. Chem. 82, 1515–1520 (2010).
Li, T., Wang, E. & Dong, S. G-quadruplex-based DNAzyme for facile colorimetric detection of thrombin. Chem. Commun. (Camb.) 3654–3656 (2008).
Li, T., Dong, S. & Wang, E. Label-free colorimetric detection of aqueous mercury ion (Hg2+) using Hg2+-modulated G-quadruplex-based DNAzymes. Anal. Chem. 81, 2144–2149 (2009).
Lu, N., Shao, C. & Deng, Z. Rational design of an optical adenosine sensor by conjugating a DNA aptamer with split DNAzyme halves. Chem. Commun. (Camb.) 6161–6163 (2008).
Lu, N., Shao, C. & Deng, Z. Colorimetric Hg2+ detection with a label-free and fully DNA-structured sensor assembly incorporating G-quadruplex halves. Analyst (Lond.) 134, 1822–1825 (2009).
Elbaz, J., Moshe, M., Shlyahovsky, B. & Willner, I. Cooperative multicomponent self-assembly of nucleic acid structures for the activation of DNAzyme cascades: a paradigm for DNA sensors and aptasensors. Chemistry 15, 3411–3418 (2009).
Constantin, T.P. et al. Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org. Lett. 10, 1561–1564 (2008).
Furutani, C., Shinomiya, K., Aoyama, Y., Yamada, K. & Sando, S. Modular blue fluorescent RNA sensors for label-free detection of target molecules. Mol. Biosyst. 6, 1569–1571 (2010).
Yoshida, W., Sode, K. & Ikebukuro, K. Aptameric enzyme subunit for biosensing based on enzymatic activity measurement. Anal. Chem. 78, 3296–3303 (2006).
Chelyapov, N. Allosteric aptamers controlling a signal amplification cascade allow visual detection of molecules at picomolar concentrations. Biochemistry 45, 2461–2466 (2006).
Stojanovic, M.N., Mitchell, T.E. & Stefanovic, D. Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 124, 3555–3561 (2002).
Penchovsky, R. & Breaker, R.R. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat. Biotechnol. 23, 1424–1433 (2005).
Stojanovic, M.N. & Stefanovic, D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 21, 1069–1074 (2003).
Kolpashchikov, D.M. & Stojanovic, M.N. Boolean control of aptamer binding states. J. Am. Chem. Soc. 127, 11348–11351 (2005).
Elbaz, J. et al. DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotechnol. 5, 417–422 (2010).A biocomputing platform with modular architecture based on a library of DNAzyme subunits, pre-designed substrates and different inputs is described.
Yoshida, W. & Yokobayashi, Y. Photonic Boolean logic gates based on DNA aptamers. Chem. Commun. 2007, 195–197 (2007).
Win, M.N. & Smolke, C.D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. USA 104, 14283–14288 (2007); erratum 106, 15514 (2009).
Win, M.N. & Smolke, C.D. Higher-order cellular information processing with synthetic RNA devices. Science 322, 456–460 (2008).
Klug, S.J., Huttenhofer, A., Kromayer, M. & Famulok, M. In vitro and in vivo characterization of novel mRNA motifs that bind special elongation factor SelB. Proc. Natl. Acad. Sci. USA 94, 6676–6681 (1997).
Choi, K.H. et al. Intracellular expression of the T-cell factor-1 RNA aptamer as an intramer. Mol. Cancer Ther. 5, 2428–2434 (2006).
Famulok, M., Blind, M. & Mayer, G. Intramers as promising new tools in functional proteomics. Chem. Biol. 8, 931–939 (2001).
Lee, H.K. et al. B-catenin regulates multiple steps of RNA metabolism as revealed by the RNA aptamer in colon cancer cells. Cancer Res. 67, 9315–9321 (2007).
Werstuck, G. & Green, M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science 282, 296–298 (1998).
Wieland, M. & Hartig, J.S. Artificial riboswitches: synthetic mRNA-based regulators of gene expression. ChemBioChem 9, 1873–1878 (2008).
Suess, B. & Weigand, J.E. Engineered riboswitches: overview, problems and trends. RNA Biol. 5, 24–29 (2008).
Win, M.N., Liang, J.C. & Smolke, C.D. Frameworks for programming biological function through RNA parts and devices. Chem. Biol. 16, 298–310 (2009).
Saito, H. & Inoue, T. Synthetic biology with RNA motifs. Int. J. Biochem. Cell Biol. 41, 398–404 (2009).
Topp, S. & Gallivan, J.P. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 5, 139–148 (2010).
Khvorova, A., Lescoute, A., Westhof, E. & Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10, 708–712 (2003).
Wieland, M. & Hartig, J.S. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Edn Engl. 47, 2604–2607 (2008).
Ogawa, A. & Maeda, M. An artificial aptazyme-based riboswitch and its cascading system in E. coli. ChemBioChem 9, 206–209 (2008).
Wieland, M., Benz, A., Klauser, B. & Hartig, J.S. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Edn Engl. 48, 2715–2718 (2009).
Wieland, M., Berschneider, B., Erlacher, M.D. & Hartig, J.S. Aptazyme-mediated regulation of 16S ribosomal RNA. Chem. Biol. 17, 236–242 (2010).
Ogawa, A. & Maeda, M. A novel label-free biosensor using an aptazyme-suppressor-tRNA conjugate and an amber mutated reporter gene. ChemBioChem 9, 2204–2208 (2008).
Berschneider, B., Wieland, M., Rubini, M. & Hartig, J.S. Small-molecule-dependent regulation of transfer RNA in bacteria. Angew. Chem. Int. Edn Engl. 48, 7564–7567 (2009). Demonstration that aminoacylation of a suppressor tRNA can be used to regulate the expression of an amber-mutated gene.
Dixon, N. et al. Reengineering orthogonally selective riboswitches. Proc. Natl. Acad. Sci. USA 107, 2830–2835 (2010).
Topp, S. & Gallivan, J.P. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 129, 6807–6811 (2007).
Sinha, J., Reyes, S.J. & Gallivan, J.P. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 6, 464–470 (2010). An application-driven approach in which bacteria are reprogrammed to move into the direction of a toxin and then to take up and metabolize this toxin.
Buskirk, A.R., Landrigan, A. & Liu, D.R. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 11, 1157–1163 (2004).
Ausländer, S., Ketzer, P. & Hartig, J.S. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 6, 807–814 (2010). The first example of use of the full-length hammerhead ribozyme to control gene expression in mammalian cells.
Chen, Y.Y., Jensen, M.C. & Smolke, C.D. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. USA 107, 8531–8536 (2010).
Ogawa, A. Rational design of artificial riboswitches based on ligand-dependent modulation of internal ribosome entry in wheat germ extract and their applications as label-free biosensors. RNA 17, 478–488 (2011).
Buratti, E. & Baralle, F.E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 24, 10505–10514 (2004).
Kim, D.S., Gusti, V., Pillai, S.G. & Gaur, R.K. An artificial riboswitch for controlling pre-mRNA splicing. RNA 11, 1667–1677 (2005).
Kim, D.S., Gusti, V., Dery, K.J. & Gaur, R.K. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol. Biol. 9, 23 (2008).
Weigand, J.E. & Suess, B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 35, 4179–4185 (2007).
Culler, S.J., Hoff, K.G. & Smolke, C.D. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255 (2010). This paper describes ligand-induced control over gene expression through regulation of splicing activity using proteins as inputs, resulting in the ability to rewire cellular networks.
Kötter, P., Weigand, J.E., Meyer, B., Entian, K.D. & Suess, B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 37, e120 (2009).
Kumar, D., An, C.I. & Yokobayashi, Y. Conditional RNA interference mediated by allosteric ribozyme. J. Am. Chem. Soc. 131, 13906–13907 (2009).
An, C.I., Trinh, V.B. & Yokobayashi, Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA 12, 710–716 (2006).
Beisel, C.L., Bayer, T.S., Hoff, K.G. & Smolke, C.D. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol. Syst. Biol. 4, 224 (2008).
Suess, B. et al. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 31, 1853–1858 (2003).
Koizumi, M., Soukup, G.A., Kerr, J.N. & Breaker, R.R. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat. Struct. Biol. 6, 1062–1071 (1999).
Piganeau, N., Thuillier, V. & Famulok, M. In vitro selection of allosteric ribozymes: theory and experimental validation. J. Mol. Biol. 312, 1177–1190 (2001).
Muranaka, N., Abe, K. & Yokobayashi, Y. Mechanism-guided library design and dual genetic selection of synthetic OFF riboswitches. ChemBioChem 10, 2375–2381 (2009).
Topp, S. & Gallivan, J.P. Random walks to synthetic riboswitches—a high-throughput selection based on cell motility. ChemBioChem 9, 210–213 (2008).
Lynch, S.A. & Gallivan, J.P. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 37, 184–192 (2009). A fast intracellular screening approach involving fluorescence-assisted cell sorting, yielding one of the strongest selected switches for gene regulation so far.
Fowler, C.C., Brown, E.D. & Li, Y.A. FACS-based approach to engineering artificial riboswitches. ChemBioChem 9, 1906–1911 (2008).
Weigand, J.E. et al. Screening for engineered neomycin riboswitches that control translation initiation. RNA 14, 89–97 (2008).
Wallis, M.G., von Ahsen, U., Schroeder, R. & Famulok, M. A novel RNA motif for neomycin recognition. Chem. Biol. 2, 543–552 (1995).
Chen, X., Denison, L., Levy, M. & Ellington, A.D. Direct selection for ribozyme cleavage activity in cells. RNA 15, 2035–2045 (2009).
Vuyisich, M. & Beal, P.A. Controlling protein activity with ligand-regulated RNA aptamers. Chem. Biol. 9, 907–913 (2002).
Acknowledgements
We apologize to authors whose work we could not cite owing to space limitations. We are grateful for funding from the Alexander von Humboldt Foundation, the North Rhine–Westphalia research school 'LIMES Chemical Biology', the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, the European Science Foundation and the European Research Council. We also thank B. Weiche for reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Vinkenborg, J., Karnowski, N. & Famulok, M. Aptamers for allosteric regulation. Nat Chem Biol 7, 519–527 (2011). https://doi.org/10.1038/nchembio.609
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.609
This article is cited by
-
Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction
Nature Communications (2023)
-
A kinetically controlled platform for ligand-oligonucleotide transduction
Nature Communications (2021)
-
Regulation of Glycine Cleavage and Detoxification by a Highly Conserved Glycine Riboswitch in Burkholderia spp.
Current Microbiology (2021)
-
Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction
Nature Communications (2020)
-
Precision immunomodulation with synthetic nucleic acid technologies
Nature Reviews Materials (2019)