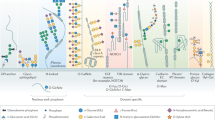
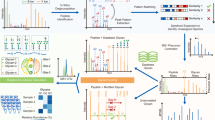
Abstract
Protein glycosylation is an important post-translational modification. It is a feature that enhances the functional diversity of proteins and influences their biological activity. A wide range of functions for glycans have been described, from structural roles to participation in molecular trafficking, self-recognition and clearance. Understanding the basis of these functions is challenging because the biosynthetic machinery that constructs glycans executes sequential and competitive steps that result in a mixture of glycosylated variants (glycoforms) for each glycoprotein. Additionally, naturally occurring glycoproteins are often present at low levels, putting pressure on the sensitivity of the analytical technologies. No universal method for the rapid and reliable identification of glycan structure is currently available; hence, research goals must dictate the best method or combination of methods. To this end, we introduce some of the major technologies routinely used for structural N- and O-glycan analysis, describing the complementary information that each provides.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dwek, R.A. Glycobiology: Toward understanding the function of sugars. Chem. Rev. 96, 683–720 (1996).
Dennis, J.W., Nabi, I.R. & Demetriou, M. Metabolism, cell surface organization, and disease. Cell 139, 1229–1241 (2009).
Freeze, H.H. & Aebi, M. Altered glycan structures: the molecular basis of congenital disorders of glycosylation. Curr. Opin. Struct. Biol. 15, 490–498 (2005).
Peracaula, R., Barrabés, S., Sarrats, A., Rudd, P.M. & de Llorens, R. Altered glycosylation in tumours focused to cancer diagnosis. Dis. Markers 25, 207–218 (2008).
Richards, M.R. & Lowary, T.L. Chemistry and biology of galactofuranose-containing polysaccharides. ChemBioChem 10, 1920–1938 (2009).
Herget, S. et al. Statistical analysis of the bacterial carbohydrate structure data base (BCSDB): Characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct. Biol. 8, 35 (2008).
Stimson, E. et al. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17, 1201–1214 (1995).
Schoenhofen, I.C. et al. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J. Biol. Chem. 281, 723–732 (2006).
Varki, A. Multiple changes in sialic acid biology during human evolution. Glycoconj. J. 26, 231–245 (2009).
Varki, A. & Marth, J.D. Oligosaccharides in vertebrate development. Semin. Dev. Biol. 6, 127–138 (1995).
Parodi, A.J. Reglucosylation of glycoproteins and quality control of glycoprotein folding in the endoplasmic reticulum of yeast cells. Biochimica et Biophysica Acta (BBA)-General Subjects 1426, 287–295 (1999).
Fukuda, M., Sasaki, H. & Fukuda, M.N. Structure and role of carbohydrate in human erythropoietin. Adv. Exp. Med. Biol. 271, 53–67 (1989).
Arnold, J.N., Wormald, M.R., Sim, R.B., Rudd, P.M. & Dwek, R.A. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25, 21–50 (2007).
Cabib, E. & Leloir, L.F. Guanosine diphosphate mannose. J. Biol. Chem. 206, 779–790 (1954).
Ohtsubo, K. & Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 (2006).
Shakin-Eshleman, S.H., Remaley, A.T., Eshleman, J.R., Wunner, W.H. & Spitalnik, S.L. N-linked glycosylation of rabies virus glycoprotein. Individual sequons differ in their glycosylation efficiencies and influence on cell surface expression. J. Biol. Chem. 267, 10690–10698 (1992).
Kornfeld, R. & Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54, 631–664 (1985).
Nilsson, I.M. & von Heijne, G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268, 5798–5801 (1993).
Spiro, R.G. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R (2002).
White, T. et al. Purification and cDNA cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J. Biol. Chem. 270, 24156–24165 (1995).
Ten Hagen, K.G., Fritz, T.A. & Tabak, L.A. All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13, 1R–16R (2003).
Hansen, J.E. et al. NetOglyc: Prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 15, 115–130 (1998).
Wandall, H.H. et al. The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology 17, 374–387 (2007).
Wittwer, A.J. et al. Effects of N-glycosylation on in vitro activity of Bowes melanoma and human colon fibroblast derived tissue plasminogen activator. Biochemistry 28, 7662–7669 (1989).
Kapitany, R.A. & Zebrowski, E.J. A high resolution PAS stain for polyacrylamide gel electrophoresis. Anal. Biochem. 56, 361–369 (1973).
Hirabayashi, J. Concept, strategy and realization of lectin-based glycan profiling. J. Biochem. 144, 139–147 (2008).
Tretter, V., Altmann, F. & Marz, L. Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha (1–3) to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199, 647–652 (1991).
Royle, L., Radcliffe, C.M., Dwek, R.A. & Rudd, P.M. (eds.) Detailed structural analysis of N-glycans released from glycoproteins in SDS-PAGE gel bands using HPLC combined with exoglycosidase array digestions. in Glycobiology Protocols 125–143 (Humana Press Inc., Totowa, NJ, 2006).
Küster, B., Wheeler, S.F., Hunter, A.P., Dwek, R.A. & Harvey, D.J. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal. Biochem. 250, 82–101 (1997).
Bhavanandan, V.P., Umemoto, J. & Davidson, E.A. Characterization of an endo-α-N-acetyl galactosaminidase from Diplococcus pneumoniae. Biochem. Biophys. Res. Commun. 70, 738–745 (1976).
Merry, A.H. et al. Recovery of intact 2-aminobenzamide-labeled O-glycans released from glycoproteins by hydrazinolysis. Anal. Biochem. 304, 91–99 (2002).
Carlson, D.M. Oligosaccharides isolated from pig submaxillary mucin. J. Biol. Chem. 241, 2984–2986 (1966).
Huang, Y.M.Y. & Novotny, M.V. Microscale nonreductive release of O-linked glycans for subsequent analysis through MALDI mass spectrometry and capillary electrophoresis. Anal. Chem. 73, 6063–6069 (2001).
Royle, L. et al. An analytical and structural database provides a strategy for sequencing O-glycans from microgram quantities of glycoproteins. Anal. Biochem. 304, 70–90 (2002).
Manzi, A. Acid hydrolysis for release of monosaccharides. in Current Protocols in Molecular Biology (eds. Ausubel, F.A., et al.) 17.16.1–17.16.11 (John Wiley and Sons, Inc., 2001).
Townsend, R.R. & Hardy, M.R. Analysis of glycoprotein oligosaccharides using high-pH anion exchange chromatography. Glycobiology 1, 139–147 (1991).
Merkle, R.K. & Popper, I. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 230, 1–15 (1994).
Schauer, R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 19, 507–514 (2009).
Hara, S., Takemori, Y., Yamaguchi, M., Nakamura, M. & Ohkura, Y. Fluorometric high-performance liquid chromatography of N-acetyl- and N-glycolylneuraminic acids and its application to their microdetermination in human and animal sera, glycoproteins, and glycolipids. Anal. Biochem. 164, 138–145 (1987).
Raju, T.S. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr. Opin. Immunol. 20, 471–478 (2008).
Hossler, P., Khattak, S.F. & Li, Z.J. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 19, 936–949 (2009).
Royle, L., Dwek, R.A. & Rudd, P.M. Determining the structure of oligosaccharides N- and O-linked to glycoproteins. in Current Protocols in Protein Science (eds. Coligan, J.E., et al.) 12.6.1–12.6.45 (John Wiley and Sons, Inc., 2006).
Kotani, N. & Takasaki, S. Analysis of 2-aminobenzamide-labeled oligosaccharides by high-pH anion-exchange chromatography with fluorometric detection. Anal. Biochem. 264, 66–73 (1998).
Domann, P.J. et al. Separation-based glycoprofiling approaches using fluorescent labels. Proteomics 7, 70–76 (2007).
Lamari, F.N., Kuhn, R. & Karamanos, N.K. Derivatization of carbohydrates for chromatographic, electrophoretic and mass spectrometric structure analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 793, 15–36 (2003).
Artemenko, N.V., Campbell, M.P. & Rudd, P.M. GlycoExtractor: A web-based interface for high throughput processing of HPLC-glycan data. J. Proteome Res. 9, 2037–2041 (2010).
Campbell, M.P., Royle, L., Radcliffe, C.M., Dwek, R.A. & Rudd, P.M. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics 24, 1214–1216 (2008).
Royle, L. et al. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem. 376, 1–12 (2008).
Knežević, A. et al. Variability, heritability and environmental determinants of human plasma N-glycome. J. Proteome Res. 8, 694–701 (2008).
Ahn, J., Bones, J., Yu, Y.-Q., Rudd, P.M. & Gilar, M. Separation of 2-aminobenzamide labeled glycans using hydrophilic interaction chromatography columns packed with 1.7 [mu]m sorbent. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878, 403–408 (2010).
Klockow, A., Michael Widmer, H., Amado, R. & Paulus, A. Capillary electrophoresis of ANTS labelled oligosaccharide ladders and complex carbohydrates. Fresenius' J. Anal. Chem. 350, 415–425 (1994).
Guttman, A. Multistructure sequencing of N-linked fetuin glycans by capillary gel electrophoresis and enzyme matrix digestion. Electrophoresis 18, 1136–1141 (1997).
Laroy, W., Contreras, R. & Callewaert, N. Glycome mapping on DNA sequencing equipment. Nat. Protoc. 1, 397–405 (2006).
Mechref, Y. & Novotny, M.V. Glycomic analysis by capillary electrophoresis-mass spectrometry. Mass Spectrom. Rev. 28, 207–222 (2009).
Kang, P., Mechref, Y. & Novotny, M.V. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun. Mass Spectrom. 22, 721–734 (2008).
Powell, A.K. & Harvey, D.J. Stabilization of sialic acids in N-linked oligosaccharides and gangliosides for analysis by positive ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 10, 1027–1032 (1996).
Wada, Y. et al. Comparison of the methods for profiling glycoprotein glycans–HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology 17, 411–422 (2007).
Cooper, H.J., Hakansson, K. & Marshall, A.G. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom. Rev. 24, 201–222 (2005).
Håkansson, K. et al. Combined electron capture and infrared multiphoton dissociation for multistage MS/MS in a Fourier transform ion cyclotron resonance mass spectrometer. Anal. Chem. 75, 3256–3262 (2003).
Wu, S.-L., Huhmer, A.F.R., Hao, Z. & Karger, B.L. On-line LCMS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J. Proteome Res. 6, 4230–4244 (2007).
Scott, N.E. et al. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, HCD and ETD-MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics published online, doi:10.1074/mcp.M000031-MCP201 (1 April 2010).
Adamson, J.T. & Hakansson, K. Electron capture dissociation of oligosaccharides ionized with alkali, alkaline earth, and transition metals. Anal. Chem. 79, 2901–2910 (2007).
Zhao, C., Xie, B., Chan, S.-Y., Costello, C.E. & O'Connor, P.B. Collisionally activated dissociation and electron capture dissociation provide complementary structural information for branched permethylated oligosaccharides. J. Am. Soc. Mass Spectrom. 19, 138–150 (2008).
Wuhrer, M., Koeleman, C.A. & Deelder, A.M. Two-dimensional HPLC separation with reverse-phase-nano-LC-MS/MS for the characterization of glycan pools after labeling with 2-aminobenzamide. in Glycomics: Methods and Protocols 1–13 (Humana Press Inc., Totowa, NJ, 2009).
Karlsson, N.G. et al. Negative ion graphitised carbon nano-liquid chromatography/mass spectrometry increases sensitivity for glycoprotein oligosaccharide analysis. Rapid Commun. Mass Spectrom. 18, 2282–2292 (2004).
Wada, Y. et al. Comparison of methods for profiling O-glycosylation. Mol. Cell. Proteomics 9, 719–727 (2010).
Larsson, J.M., Karlsson, H., Sjovall, H. & Hansson, G.C. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19, 756–766 (2009).
Agard, N.J. & Bertozzi, C.R. Chemical approaches to perturb, profile, and perceive glycans. Acc. Chem. Res. 42, 788–797 (2009).
Alley, W.R., Madera, M., Mechref, Y. & Novotny, M.V. Chip-based reversed-phase liquid chromatography mass spectrometry of permethylated N-linked glycans: A potential methodology for cancer-biomarker discovery. Anal. Chem. 82, 5095–5106 (2010).
Chu, C.S. et al. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics 9, 1939–1951 (2009).
Wormald, M.R. et al. Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev. 102, 371–386 (2002).
Forsberg, L.S. & Carlson, R.W. Structural characterization of the primary O-antigenic polysaccharide of the Rhizobium leguminosarum 3841 lipopolysaccharide and identification of a new 3-acetimidoylamino-3-deoxyhexuronic acid glycosyl component: A unique O-methylated glycan of uniform size, containing 6-deoxy-3-O-methyl-D-talose, N-Acetylquinovosamine, and rhizoaminuronic acid (3-acetimidoylamino-3-deoxy-D-gluco-hexuronic acid). J. Biol. Chem. 283, 16037–16050 (2008).
Vliegenthart, J.F.G. Introduction to NMR spectroscopy of carbohydrates. in NMR Spectroscopy and Computer Modeling of Carbohydrates 1–19 (American Chemical Society, 2006).
Bubb, W.A. NMR spectroscopy in the study of carbohydrates: Characterizing the structural complexity. Concepts in Magnetic Resonance Part A 19A, 1–19 (2003).
Maguire, Y.C., Chuang, I.L., Zhang, S. & Gershenfeld, S. Ultra-small-sample molecular structure detection using microslot waveguide nuclear spin resonance. Proc. Natl. Acad. Sci. USA 104, 9198–9203 (2007).
Manzi, A.E. et al. Exploring the glycan repertoire of genetically modified mice by isolation and profiling of the major glycan classes and nano-NMR analysis of glycan mixtures. Glycobiology 10, 669–689 (2000).
Lütteke, T. et al. GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology 16, 71R–81R (2006).
Jansson, P.-E., Stenutz, R. & Widmalm, G. Sequence determination of oligosaccharides and regular polysaccharides using NMR spectroscopy and a novel Web-based version of the computer program casper. Carbohydr. Res. 341, 1003–1010 (2006).
Calvano, C.D., Zambonin, C.G. & Jensen, O.N. Assessment of lectin and HILIC based enrichment protocols for characterization of serum glycoproteins by mass spectrometry. J. Proteomics 71, 304–317 (2008).
Narimatsu, H. et al. A strategy for discovery of cancer glyco-biomarkers in serum using newly developed technologies for glycoproteomics. FEBS J. 277, 95–105 (2010).
Wada, Y., Tajiri, M. & Yoshida, S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal. Chem. 76, 6560–6565 (2004).
Wohlgemuth, J., Karas, M., Jiang, W., Hendriks, R. & Andrecht, S. Enhanced glyco-profiling by specific glycopeptide enrichment and complementary monolithic nano-LC (ZIC-HILIC/RP18e)/ESI-MS analysis. J. Sep. Sci. 33, 880–890 (2010).
Alley, W.R. Jr., Mechref, Y. & Novotny, M.V. Use of activated graphitized carbon chips for liquid chromatography/mass spectrometric and tandem mass spectrometric analysis of tryptic glycopeptides. Rapid Commun. Mass Spectrom. 23, 495–505 (2009).
Jiang, H., Wu, S.L., Karger, B.L. & Hancock, W.S. Characterization of the glycosylation occupancy and the active site in the follow-on protein therapeutic: TNK-tissue plasminogen activator. Anal. Chem. 82, 6154–6162 (2010).
Wuhrer, M., Boer, A.R.d. & Deelder, A.M. Structural glycomics using hydrophilic interaction chromatography (HILIC) with mass spectrometry. Mass Spectrom. Rev. 28, 192–206 (2009).
Comelli, E.M. et al. A focused microarray approach to functional glycomics: transcriptional regulation of the glycome. Glycobiology 16, 117–131 (2006).
Taylor, M.E. & Drickamer, K. Structural insights into what glycan arrays tell us about how glycan-binding proteins interact with their ligands. Glycobiology 19, 1155–1162 (2009).
Horlacher, T. et al. Determination of carbohydrate-binding preferences of human galectins with carbohydrate microarrays. ChemBioChem 11, 1563–1573 (2010).
Kiessling, L.L. & Splain, R.A. Chemical approaches to glycobiology. Annu. Rev. Biochem. 79, 619–653 (2010).
Laughlin, S.T. & Bertozzi, C.R. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat. Protoc. 2, 2930–2944 (2007).
Jervis, A.J. et al. Characterisation of N-linked protein glycosylation in Helicobacter pullorum. J. Bacteriol. published online, doi:10.1128/JB.00211–10 (25 June 2010).
Svarovsky, S.A. & Joshi, L. Biocombinatorial selection of carbohydrate binding agents of therapeutic significance. Curr. Drug Discov. Technol. 5, 20–28 (2008).
Harvey, D.J. et al. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics 9, 3796–3801 (2009).
Acknowledgements
K.M. is supported by Science Foundation Ireland, grant number 08/SRC/B1393. J.B. acknowledges the European Union FP6 GLYFDIS research program, grant reference 037661 for funding. J.J.K. acknowledges the Chief Scientist Office of the Scottish Government, the Royal Society and the European Union framework program 6 EUROSPAN project (contract number LSHG-CT-2006-018947) for funding. The authors are grateful to M. Campbell, W. Struwe, T. Tharmalingam and J. Abrahams for critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Mariño, K., Bones, J., Kattla, J. et al. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol 6, 713–723 (2010). https://doi.org/10.1038/nchembio.437
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.437
This article is cited by
-
N-glycans from serum IgG and total serum glycoproteins specific for endometriosis
Scientific Reports (2023)
-
Advances in single-molecule junctions as tools for chemical and biochemical analysis
Nature Chemistry (2023)
-
A universal UHPLC-CAD platform for the quantification of polysaccharide antigens
Scientific Reports (2023)
-
N-Linked Glycoproteome Analysis of Diosorea alata Tuber Shows Atypical Glycosylation and Indicates Central Role of Glycosylated Proteins in Tuber Maturation
The Protein Journal (2023)
-
Comparative study for analysis of carbohydrates in biological samples
Analytical and Bioanalytical Chemistry (2022)