Abstract
Bisphosphonates are potent inhibitors of farnesyl pyrophosphate synthase (FPPS) and are highly efficacious in the treatment of bone diseases such as osteoporosis, Paget's disease and tumor-induced osteolysis. In addition, the potential for direct antitumor effects has been postulated on the basis of in vitro and in vivo studies and has recently been demonstrated clinically in early breast cancer patients treated with the potent bisphosphonate zoledronic acid. However, the high affinity of bisphosphonates for bone mineral seems suboptimal for the direct treatment of soft-tissue tumors. Here we report the discovery of the first potent non-bisphosphonate FPPS inhibitors. These new inhibitors bind to a previously unknown allosteric site on FPPS, which was identified by fragment-based approaches using NMR and X-ray crystallography. This allosteric and druggable pocket allows the development of a new generation of FPPS inhibitors that are optimized for direct antitumor effects in soft tissue.
Similar content being viewed by others
Main
Nitrogen-containing bisphosphonates block excessive bone resorption by inhibiting farnesyl pyrophosphate synthase (FPPS) in osteoclasts, thus inhibiting the mevalonate pathway, blocking the prenylation of GTPase signaling proteins and causing osteoclast apoptosis1. In contrast to common lipophilic drugs, bisphosphonates are highly charged, water-soluble molecules that can enter only those cells that show fluid-phase endocytosis, such as osteoclasts and macrophages. Because of their high affinity for bone mineral, they rapidly accumulate at sites of active bone turnover, their desired site of action. This makes them highly suitable as drugs to treat bone diseases.
The potential of bisphosphonates as antitumor agents has been suggested by several in vitro and in vivo preclinical studies (reviewed in ref. 2). A recent report of a significant reduction of disease progression in premenopausal breast cancer patients treated with endocrine therapy and zoledronic acid3 has raised great interest in these antitumor properties. Data suggesting that statins—which inhibit the mevalonate pathway by blocking HMG-CoA reductase upstream of FPPS—also have antitumor properties corroborate the importance of the mevalonate pathway as a clinically relevant antitumor target4.
On reaching the systemic circulation, approximately half of the bisphosphonate dose is rapidly adsorbed to bone, and the remainder is rapidly excreted unchanged via the kidney. Consequently, blood levels and soft-tissue exposure are very low soon after administration. In combination with the low permeability of these drugs into tumor cells and other non-endocytic cells, the pharmacokinetic properties of bisphosphonates are not optimal for antitumor treatment, and FPPS inhibitors with optimized pharmacokinetic properties would be expected to have even higher efficacy5. Several approaches have been taken to optimize the pharmacokinetic properties of bisphosphonates, such as reducing bone affinity with deoxy-bisphosphonates6 or enhancing cellular uptake by increasing side chain lipophilicity7. Although these approaches show some promise in vitro and in animal models, the physicochemical properties of the resulting molecules are still dominated by the bisphosphonate moiety. A completely new chemical class of FPPS inhibitors without inherent bone affinity could revolutionize the field and open up new opportunities for more effective antitumor treatment. In spite of decades of research, however, non-bisphosphonate FPPS inhibitors have never been identified, and it seemed plausible that they would never be identified, as bisphosphonates inhibit FPPS by mimicking one of the highly polar substrates, dimethylallyl pyrophosphate (DMAPP, Scheme 1). The requirement for a bisphosphonate moiety specifically has been substantiated by the synthesis of phosphonocarboxylate analogs of risedronate, which were shown to lack inhibitory potency8. Here we describe the discovery of an allosteric binding pocket that favors a radically different pharmacophore and allows the inhibition of FPPS by new and potent non-bisphosphonate chemotypes.
Results
Fragment-based screening
New inhibitors of FPPS were identified by fragment-based approaches9,10,11. We started by screening a small library of 400 fragments for binding affinity to human recombinant FPPS using NMR spectroscopy, a biophysical technique well suited for this purpose because of its ability to robustly detect even weak binding interactions (Fig. 1 and Supplementary Table 1). Indeed, the identified hits, among them 1, 2, 3 and 4 (Scheme 2), bound only very weakly to FPPS, with dissociation constants in the millimolar range. Those fragments would not have been found in a high-throughput biochemical assay. Although weak binders, they were still interesting starting points for optimization because of their small molecular size. In the course of characterizing these fragment hits, we performed competition studies using zoledronic acid as a high-affinity competitor. To our surprise, most fragment hits were not competitive with zoledronic acid but bound to FPPS independent of or even synergistically with zoledronic acid or other bisphosphonates (Fig. 1c). This indicated that the identified fragments bound at a binding site that was different from the DMAPP substrate site, the binding site of all known bisphosphonates12,13.
NMR screening was carried out using T1ρ relaxation experiments with 10 ms (black) and 100 ms (red) relaxation period and using waterLOGSY experiments (blue). (a) Mixture of 200 μM fragments without FPPS. (b) Mixture of fragments with 10 μM FPPS. (c) Mixture of fragments with 10 μM FPPS and 80 μM zoledronic acid (ZOL). The resonances of 4 are indicated on top. Note the lack of competition between 4 and zoledronic acid.
Discovery of an allosteric binding pocket
The binding site was precisely determined by X-ray crystallography. Crystal structures were obtained for all four fragment hits shown in Scheme 2. One of them, 4, was crystallized as a ternary complex with zoledronic acid, whereas the other three were crystallized as binary complexes. In each of these four structures, the fragment was bound outside the active site, in an allosteric pocket near the C terminus of the enzyme and adjacent to the isopentenyl pyrophosphate (IPP) binding site. This pocket is remote from the dimer interface, and there are thus two identical pockets per homodimer (Fig. 2a, electron density maps in Supplementary Fig. 1). For all binary complexes, FPPS was found to adopt the open conformation, whereas the ternary complex with 4 and zoledronic acid was in the partially closed state (for a detailed discussion of the open, partially closed and fully closed conformations, see ref. 12). The existence and nature of this allosteric pocket was previously unknown. The pocket is mainly lined by helices αC, αG, αH and the C-terminal helix αJ. In addition, a few residues from the αB-αC loop, the αH-αI loop, the N-terminal helix αA and the C-terminal loop are also contributing in some of the complexes. The pocket comprises a hydrophobic base and rear side, involving the side chains of Phe206, Phe239, Leu344, Ile348 and Tyr10. In sharp contrast, the opposite side comprises several positively charged side chains (Lys57, Arg60, Lys347) as well as polar side chains (Asn59, Thr63). The exact size and shape of the pocket is affected by the conformational transition from the open state to the partially closed state, which brings αH and αJ closer to αC and αG. The pocket is smallest in the partially closed state, as observed with 4 and zoledronic acid, and remains exposed to the solvent region because the basic carboxyterminal tail of the enzyme (Lys350-Arg351-Arg352-Lys353) is unable to clamp down and remains disordered when the pocket is occupied.
(a) Overall view of the FPPS homodimer, in the open (orange ribbon) and closed (gray ribbon) conformations. Zoledronic acid is shown as blue sticks, IPP is shown as red sticks and the newly identified allosteric pocket is shown as a green surface. (b) 4 in the allosteric binding pocket. The bound phosphate ion occupies the position of the β-phosphate in IPP. (c) Details of the interactions of 4 in the FPPS allosteric pocket. (d) Superposition of four fragment structures. (e) Details of the interactions of 2 in the FPPS allosteric pocket. (f) Structure of 8 (magenta), superimposed with its enantiomer, 4 (green). Both structures were solved as a complex with zoledronic acid. (g) Structure of 9. (h) Structure of 10 (magenta) and superposition with 9 (light blue).
4 is bound with its benzyloxycarbonyl group deeply buried within the hydrophobic lower region of the pocket, whereas its indoline moiety is more exposed, pointing toward the αH-αI loop and making contacts with Gln242, Leu246 and Val254 (Fig. 2b,c). The smaller fragments 1, 2 and 3 are all bound within the confines of the hydrophobic base of the pocket. Their binding mode appears to be dictated by the strong electrostatic interactions between the compounds' carboxylic groups and the side chains of Arg60 and Lys57 (Fig. 2d). In addition, a short hydrogen-bonded contact between the carboxylic group of the fragment and Asn59 Nδ1 is consistently observed in these three complexes. As a result of these dominant electrostatic and/or hydrogen-bonded interactions, the benzothiophene moieties of 1 and 2 adopt different orientations (Fig. 2e), while still engaging in hydrophobic and/or aromatic interactions with Phe206, Phe239, Leu344 and the alkyl portion of the Lys347 side chain. The different substitution pattern and orientations of the benzothiophenes 1 and 2 lead to small induced-fit changes of the Phe239, Leu344 and Lys347 side chains. The orientation of the benzofurane ring of 3 is again at variance with the two benzothiophenes 1 and 2, as a result of the presence of a 6-methoxy instead of 5-chloro substituent on the benzyl ring (Fig. 2d). The carboxylic group of the fragment is engaged in similar electrostatic interactions with Arg60, Lys57 and Asn59, whereas the 6-methoxy group is located deep in the base of the pocket, within hydrogen-bonding distance of Thr63 Oγ1. As with the other fragments, hydrophobic and/or aromatic interactions with Phe206, Phe239 and the alkyl portion of Lys347 are also observed. In contrast to the DMAPP substrate pocket, which was previously the only known binding pocket for FPPS inhibitors, this allosteric pocket is not Mg2+ dependent; indeed, all fragments bind in the presence or absence of Mg2+.
Fragment optimization
To see whether the allosteric pocket is druggable and functional, we assembled a small library of 40 compounds from the Novartis compound inventory, on the basis of similarity with the identified fragment hits and coarse pharmacophore criteria. As biochemical assays were unreliable because of the low affinities, we used NMR as a biophysical assay to identify compounds with higher binding affinity. Using 2 as a reporter ligand14, we identified 5, which has about a tenfold higher binding affinity. Assembly and screening of a second focused library, this time using 5 as an NMR reporter ligand, identified 6. This optimized fragment showed reproducible enzyme inhibition with a half-maximal inhibitory concentration (IC50) of 12 μM, in an assay measuring product formation by either LC-MS15 or scintillation proximity16. The evidence for enzyme inhibition by optimized fragments was very encouraging because it proved the functional relevance of the newly identified allosteric pocket and showed that ligands for this pocket are indeed effective inhibitors. By analyzing the available crystal structures, we noticed that the binding site was larger than what the current fragments could fill, and so we synthesized naphthothiophenes such as 7 (see Supplementary Methods), based on the merged structure-activity relationships (SARs) of benzothiophenes. 7 had an IC50 of 7 μM in the above-mentioned biochemical assays.
In the crystal structure of the original fragment hit 4 (Fig. 2c), we observed that the carboxylic acid functionality was not pointing in the appropriate direction to interact with Arg60, in contrast to the benzothiophene and benzofurane series, implying that the other enantiomer, 8, might bind with higher affinity. Indeed, 8 showed 20-fold increased binding affinity, and we therefore decided to solve its crystal structure, as a ternary complex with zoledronic acid. To our surprise, we found that 8 was binding upside down relative to 4 (Fig. 2f). The indoline moiety of 8, rather than its benzyloxycarbonyl group, now filled the same region of the pocket as the benzothiophenes and benzofurane fragments, with its 2-carboxylic group engaged in strong electrostatic and hydrogen-bonded interactions with Arg60, Lys57 and Asn59. We therefore assembled and screened a new focused library and identified the 2-carboxyindole derivatives 9 and 10 (Fig. 2g,h). These optimized fragments were active in the scintillation proximity and LC-MS assays with IC50 values of 2 μM and 6 μM, respectively. 10 also showed activity in a cellular plasma membrane translocation assay17 in which localization of enhanced GFP (eGFP), tagged with the prenylation motif of human H-Ras, is monitored by imaging techniques. Prenylated eGFP is localized mainly to the plasma membrane, as seen in the DMSO control, whereas inhibition of prenylation by 10 leads to cytoplasmic and nuclear localization of the eGFP reporter (Fig. 3). At higher concentrations, these compounds induce apoptosis, as seen by increased nuclear density and condensed small nuclei.
(b) Images of U-2 OS cells expressing the eGFP reporter for prenylation. The cells were treated with 0.5% (v/v) DMSO (negative control), 5 μM zoledronic acid (positive control) or 50 μM 10 for 48 h.
The crystal structures of 9 and 10 were solved (Fig. 2g,h). Both compounds were found to adopt similar binding modes, with the 3-carboxymethyl substituent interacting with Arg60, Lys57 and Asn59, and the chlorinated indole system contributing hydrophobic and/or aromatic interactions with Leu344, Tyr10, Phe206, Phe239 and Lys347. In the FPPS complex with 10 (Fig. 2h), the 5-chloro substituent is involved in a short, favorable contact to the Ser205 carbonyl oxygen. The indole ring of 9 is not substituted in position 5 and is therefore able to penetrate more deeply into the pocket. For both compounds, the carboxylate group in position 2 is likely to contribute additional electrostatic interactions, in particular with Lys350 and the following three basic residues at the carboxy terminus of the enzyme, although this part of the structure is not seen in the X-ray analysis, owing to disorder.
With the crystal structures as well as the biochemical and cellular assay data for 7, 8 and 10 in hand, we attempted to synthesize compounds that would combine all useful features by merging the SAR of these fragment series. This resulted in 11 and 12, new non-bisphosphonate FPPS inhibitors with IC50 values of 200 nM and 80 nM, respectively. Notably, the potencies of these compounds in biochemical assays are similar to those of the most potent bisphosphonate, zoledronic acid (IC50 = 130 nM), and similar to or higher than those of all other bisphosphonates used clinically, such as risedronate, ibandronate, alendronate and pamidronate (IC50 = 150 nM, 340 nM, 850 nM, 950 nM, respectively), when measured without preincubation18. The IC50 of 11 did not change after 30 min preincubation with FPPS, FPPS and GPP, or FPPS and IPP (Supplementary Fig. 2). The high potencies of our optimized compounds show that the newly discovered allosteric pocket is highly druggable and of significant pharmaceutical relevance, as it allows the discovery of highly potent nonbisphosphonate FPPS inhibitors.
Mechanism of allosteric inhibition
The mechanism of inhibition by ligands binding to the allosteric pocket involves three aspects. First, the C-terminal tail of FPPS—which, for catalysis, has to adopt the fully closed conformation upon binding of the second substrate, IPP12—is sterically hindered from closing by allosteric ligands (Fig. 4 and Supplementary Fig. 3). Second, there would be electrostatic repulsion between negatively charged allosteric ligands and IPP, which come close together (Fig. 2a). Third, although—at least for 4 and 8—allosteric ligands can bind to the closed as well as the open conformation, binding of allosteric ligands to either conformation probably disturbs the dynamics of opening and closing of the enzyme during catalysis.
(a) Size of the allosteric pocket in the open FPPS conformation. (b) Size of the allosteric pocket in the fully closed conformation. The three C-terminal FPPS residues that fully close the active site in the IPP complex and their surface contributions are shown in green. Note the potential steric clash of a bound fragment with the three C-terminal FPPS residues in the fully closed conformation and the potential close proximity of negative charges in the IPP complex.
Lead characterization
In order to confirm the high binding affinity of 11 in an independent biophysical assay, we performed isothermal titration calorimetry (ITC). A 1:1 binding stoichiometry was found with each subunit in the FPPS homodimer. The binding enthalpy and entropy in this enthalpy-driven binding event were −12.9 kcal mol−1 and −12.5 cal mol−1 K−1, respectively, and the resulting KD was 180 nM, in good agreement with the measured IC50 value in the enzymatic assay (Supplementary Fig. 4). The KD values determined by ITC for zoledronic acid were 180 nM (in-house, data not shown) and 15 nM (ref. 13). The crystal structure of 11 shows that this compound adopts a binding mode that is most similar to that of the original fragment hit 1 (Supplementary Fig. 5). The naphthyl moiety of 11 superimposes onto the benzothiophene ring system of 1, whereas its five-membered ring occupies the position of the 5-chloro substituent. The carboxymethyl substituents of 11 and 1 come from different positions, but their carboxylic groups bind next to each other, between the Arg60 and Lys57 side chains. The carboxylic group in position 2 of 11 appears to be pointing toward the solvent region but contributes significantly to binding potency, presumably through additional electrostatic interactions with the disordered carboxy-terminal tail of the enzyme. Notably, the ligand efficiency and binding efficiency index have significantly increased in the course of optimizing the initial fragment hits to the lead compounds19.
In the Novartis preclinical profiling panel, 11 showed low transcellular permeability (logP = −5.6 for gastrointestinal absorption) in a parallel artificial membrane permeability assay (PAMPA)20, medium-to-low permeability (Papp A–B = 1.49 × 106 cm sec−1, Papp B–A = 2.07 × 106 cm sec−1) in a Caco-2 cellular assay, no hERG channel inhibition at 30 μM and no IC50 values below 10 μM in an in vitro safety pharmacology panel of over 50 receptors21. None of the compounds 1–12 showed measurable affinity to hydroxyapatite or bone powder in an NMR-based binding assay22 (Supplementary Fig. 6). Although the drug-like properties of 11 and 12, such as cellular permeability, need to be improved, their new chemotype, in vitro safety data and lack of bone affinity indicate the high potential of non-bisphosphonate FPPS inhibitors binding at the allosteric pocket. Fragment-based screening provided a head start with respect to the full-deck high-throughput screen, which was completed after the discovery of 11 and did not result in viable hits with IC50 < 5 μM.
Discussion
11 and 12 are the first reported potent non-bisphosphonate FPPS inhibitors. Their discovery by fragment-based screening was facilitated by the unexpected discovery of a druggable allosteric binding pocket near the C terminus of the enzyme. The natural function of this allosteric pocket is still elusive. On the basis of its preferred binding of lipophilic molecules containing a negative charge, we suspected a regulatory role with negative feedback by downstream products, but neither cholesterol nor a selection of bile acids inhibited the enzyme. Nucleotides such as ATP, GTP or their analogs did not inhibit the enzyme either. The elucidation of the natural function of this pocket will be an interesting task for the future.
The discovery of non-bisphosphonate FPPS inhibitors is an example of the successes and surprises encountered when applying modern drug discovery technologies to well-known targets. After decades of futile attempts to modify the bisphosphonate moiety in order to identify new classes of FPPS inhibitors with different pharmacokinetic properties, the discovery of the allosteric pocket and potent non-bisphosphonate inhibitors such as 11 and 12 is a breakthrough in the ongoing search for new antitumor drugs based on FPPS inhibition. With these new allosteric inhibitors, FPPS inhibition is not associated with high affinity to bone mineral. This allows the application of these inhibitors outside the field of bone diseases. FPPS is an essential enzyme in the mevalonate pathway and just a few steps downstream of HMG-CoA reductase, the molecular target of statins. This suggests that non-bisphosphonate FPPS inhibitors could serve as cholesterol-lowering agents in conjunction with or as an alternative to statins. In fact, lower serum cholesterol levels have been reported in myeloma patients treated with zoledronic acid23, and weaker nonsignificant effects on cholesterol levels have been observed following administration of pamidronate to patients with Paget's disease of bone24. These effects may be more pronounced for non-bisphosphonate inhibitors, owing to their different tissue distribution profile.
The mevalonate pathway has also been implicated in other diseases such as Alzheimer's disease, and a variety of clinical trials to explore the benefits of statins are ongoing25,26. Bisphosphonates have been shown to be efficacious against parasitic protozoa causing leishmaniasis, malaria, cryptosporidiosis and Chagas's disease (reviewed in ref. 27), but again non-bisphosphonate FPPS inhibitors may be better suited for such applications, owing to their lack of affinity for bone. In another study, statins and bisphosphonates together have been reported to be efficacious in a mouse model of progeria28. Finally, viperin, a negative regulator of viral diseases such as hepatitis C, is regulated by FPPS activity, and bisphosphonates have been shown to decrease viral activity29. Excessive prenylation of target proteins appears to be a causative factor in various diseases. For all of these mevalonate pathway-dependent non-bone diseases, inhibition of the mevalonate pathway at a different position and possibly in conjunction with statins is a promising strategy. Allosteric, non-bisphosphonate inhibitors of FPPS, as reported here, open up new opportunities for the experimental validation of this therapeutic approach.
Methods
Cloning, expression and purification of human FPPS.
Human FPPS (residues 6–353 with an N-terminal His6 tag followed by a PreScission protease cleavage site) was overexpressed in Escherichia coli BL21 (DE3) Tuner cells (Novagen). The soluble fraction of His-tagged FPPS was purified from the cell lysate by anion exchange (Q-Sepharose HP), hydrophobic (Phenyl Sepharose HP) and metal chelation (HiTrap) chromatography. The N-terminal His6 tag was then removed with the PreScission protease, and the reaction products were separated by metal chelation chromatography. His tag–free FPPS was further purified by size-exclusion chromatography (Superdex-200), concentrated by ultrafiltration to 16 mg ml−1 and stored at −80 °C.
NMR spectroscopy.
Fragment-based screening using NMR spectroscopy was carried out at 23 °C on a Bruker AV600 MHz spectrometer at a 1H resonance frequency of 600 MHz. A library of 400 fragments, in mixtures of 8, was screened for interactions with FPPS using T1ρ and the waterLOGSY NMR methodology30. The FPPS concentration was 10 μM, and each fragment was at 200 μM concentration. The buffer consisted of 25 mM d-Tris, pH 7.4, 2 mM d-DTT and 25 mM NaCl. Competition experiments with zoledronate were carried out with 80 μM zoledronate, using T1ρ and waterLOGSY experiments for the readout. After initial fragment screening, focused follow-up libraries of around 40 compounds each were tested by reporter screening, for which the displacement by follow-up compounds of a previously identified fragment was monitored by T1ρ spectra. The first round of optimization used 2 as reporter ligand, and further rounds used 5 and 6 as reporter ligands. 6 was potent enough to show reproducible activity in a biochemical assay, and the biochemical assay was then used for further optimization.
Enzymatic assay.
Biochemical enzymatic assays measured the inhibition of product formation either by LC-MS analysis of farnesyl pyrophosphate (FPP) production15 or by a scintillation proximity assay using 3H-labeled isopentenyl pyrophosphate and lipid-coated 'flashplates' with embedded scintillant16. All measurements were carried out as 8-point dose response curves and are reported as the average of at least four independent measurements. 11 was measured 21 times, with average IC50 = 0.2 ± 0.06 μM and a mean Hill coefficient of 1.04.
Isothermal titration microcalorimetry (ITC).
ITC measurements were performed on a VP-ITC calorimeter (Microcal, Inc.). The protein was dialyzed extensively against the buffer solution (50 mM HEPES, pH 7.5, 25 mM NaCl, 2 mM DTT), and the ligand was dissolved in the last dialysate. Calorimetric titrations were carried out at 25 °C. A typical titration involved 25–30 injections of each ligand (4–10 μl aliquot per injection) at 3-min intervals into the sample cell containing the protein solution. The titration cell was stirred continuously at 310 r.p.m. The heat of the ligand dilution in the buffer alone was subtracted from the titration data for each drug. The data were analyzed to determine binding stoichiometry (N), binding affinity (Kb) and thermodynamic parameters of the reaction using Origin software (OriginLab Corp.).
Translocation assay.
A plasma membrane translocation assay was carried out as described in Simonen et al.17. U-2 OS cells, stably transfected with an eGFP reporter construct for prenylation, were incubated with 50 μM 10 and 0.5% DMSO or with 0.5% DMSO only for 48 h. Then the cells were fixed, and images were acquired with IN Cell analyzer 2000.
Crystallization.
All crystals were grown by the method of vapor diffusion in hanging drops at 19 °C, from a crystallization buffer consisting of 1.2 M NaH2PO4/K2HPO4, pH 5.3, and 25% (v/v) glycerol. The compounds were dissolved in the crystallization buffer, and crystallization drops were set up by mixing equal volumes of this solution and of an FPPS stock solution in 10 mM Tris-HCl, pH 7.4, 25 mM NaCl, 5 mM TCEP, 5 mM MgCl2. 1, 2, 3 and 4 were cocrystallized at a concentration of 20 mM, whereas lower concentrations were used for all other compounds (5 mM for 8 and 10; 2.4 mM for 11; 1.2 mM for 9 and 7; 1 mM zoledronic acid was added in the case of 4 and 8). The FPPS concentration was 16 mg ml−1 for all complexes except 10, 8 and 11, which were crystallized at 26–32 mg ml−1.
X-ray data collection, structure determination and refinement.
All crystals were mounted in cryoloops and flash frozen for data collection at 95–100 K. Only one crystal was used for each dataset. The FPPS complexes with 7, 9 and 11 were measured with a Saturn92 CCD detector mounted on a FR-E rotating anode generator using 1.54178-Å radiation. All other crystals were measured with a MARCCD detector using synchrotron radiation (Swiss Light Source, beamline XS06A, wavelength 0.97934 Å). Diffraction data were processed with HKL200031 or with XDS32. All crystals were in space group P41212 with one FPPS subunit per asymmetric unit. The structures of the complexes with 4 and 8 were determined with APRV33, using an initial rigid-body refinement of the previously solved FPPS complex with zoledronic acid12, and then manual fitting of the compound into difference electron density and full refinement with CNX34 and O35. All other complexes were solved in a similar way using the structure of unliganded FPPS12 as a starting model. For 1, 3, 9 and 10, two molecules of compound were observed—the first one in the allosteric binding site, the second, more weakly bound one between two FPPS molecules in the crystal ('crystal packing site'). Our inspection of this crystal packing site (Supplementary Fig. 7), the significantly higher B factors of some ligands there (Supplementary Table 2), the fact that only some ligands were found in the site and the 1:1 stoichiometry of ITC data with 11 all indicate that this site is a crystal artifact without biological significance. X-ray data collection and refinement statistics are summarized in Supplementary Table 2.
Chemical synthesis.
The synthesis of non-bisphosphonate FPPS inhibitors is described in Supplementary Methods.
Accession codes.
The FPPS complex with zoledronic acid and IPP was deposited as part of a previous study12 under Protein Data Bank accession code 2F8Z. The coordinates of the FPPS complexes with 1, 2, 3, 4, 7, 8, 9, 10 and 11 have been deposited under accession codes 3N1V, 3N1W, 3N3L, 3N45, 3N49, 3N46, 3N5H, 3N5J and 3N6K, respectively.
Additional information
Supplementary information, chemical compound and chemical probe information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/. Correspondence and requests for materials should be addressed to W.J.

(a) Scheme of the FPPS reactions, showing the ligation of dimethylallyl pyrophosphate (DMAPP) and isopentenyl pyrophosphate (IPP) to geranyl pyrophosphate (GPP) and of GPP and IPP to farnesyl pyrophosphate (FPP). (b) The most potent clinically used bisphosphonates: zoledronate, ibandronate and risedronate.

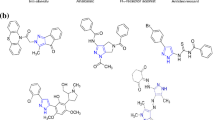
Chemical structures of fragment hits and optimized inhibitors.
References
Rogers, M.J. New insights into the molecular mechanisms of action of bisphosphonates. Curr. Pharm. Des. 9, 2643–2658 (2003).
Stresing, V., Daubine, F., Benzaid, I., Monkkonen, H. & Clezardin, P. Bisphosphonates in cancer therapy. Cancer Lett. 257, 16–35 (2007).
Gnant, M. et al. Endocrine therapy plus zoledronic acid in premenopausal breast cancer. N. Engl. J. Med. 360, 679–691 (2009).
Poynter, J.N. et al. Statins and the risk of colorectal cancer. N. Engl. J. Med. 352, 2184–2192 (2005).
Fournier, P.G. et al. Lowering bone mineral affinity of bisphosphonates as a therapeutic strategy to optimize skeletal tumor growth inhibition in vivo. Cancer Res. 68, 8945–8953 (2008).
Simoni, D. et al. Design, synthesis, and biological evaluation of novel aminobisphosphonates possessing an in vivo antitumor activity through a gammadelta-T lymphocytes-mediated activation mechanism. J. Med. Chem. 51, 6800–6807 (2008).
Zhang, Y. et al. Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation. J. Am. Chem. Soc. 131, 5153–5162 (2009).
Marma, M.S. et al. Synthesis and biological evaluation of alpha-halogenated bisphosphonate and phosphonocarboxylate analogues of risedronate. J. Med. Chem. 50, 5967–5975 (2007).
Hajduk, P.J. & Greer, J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 6, 211–219 (2007).
Jahnke, W. & Erlanson, D.A. (eds.). Fragment-based Approaches in Drug Discovery (Wiley-VCH, 2006).
Murray, C.W. & Rees, D.C. The rise of fragment-based drug discovery. Nature Chemistry 1, 187–192 (2009).
Rondeau, J.M. et al. Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs. ChemMedChem 1, 267–273 (2006).
Kavanagh, K.L. et al. The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc. Natl. Acad. Sci. USA 103, 7829–7834 (2006).
Jahnke, W. et al. NMR reporter screening for the detection of high-affinity ligands. Angew. Chem. Int. Edn Engl. 41, 3420–3423 (2002).
Roddy, T.P. et al. Mass spectrometric techniques for label-free high-throughput screening in drug discovery. Anal. Chem. 79, 8207–8213 (2007).
Glickman, J.F. & Schmid, A. Farnesyl pyrophosphate synthase: real-time kinetics and inhibition by nitrogen-containing bisphosphonates in a scintillation assay. Assay Drug Dev. Technol. 5, 205–214 (2007).
Simonen, M. et al. High-content assay to study protein prenylation. J. Biomol. Screen. 13, 456–467 (2008).
Dunford, J.E. et al. Structure-activity relationships among the nitrogen containing bisphosphonates in clinical use and other analogues: time-dependent inhibition of human farnesyl pyrophosphate synthase. J. Med. Chem. 51, 2187–2195 (2008).
Abad-Zapatero, C. & Metz, J.T. Ligand efficiency indices as guideposts for drug discovery. Drug Discov. Today 10, 464–469 (2005).
Wohnsland, F. & Faller, B. High-throughput permeability pH profile and high-throughput alkane/water log P with artificial membranes. J. Med. Chem. 44, 923–930 (2001).
Faller, B. et al. High-throughput in vitro profiling assays: lessons learnt from experiences at Novartis. Expert Opin. Drug Metab. Toxicol. 2, 823–833 (2006).
Jahnke, W. & Henry, C. An in vitro assay to measure targeted drug delivery to bone mineral. ChemMedChem 5, 770–776 (2010).
Gozzetti, A. et al. The effects of zoledronic acid on serum lipids in multiple myeloma patients. Calcif. Tissue Int. 82, 258–262 (2008).
Montagnani, A. et al. Changes in serum HDL and LDL cholesterol in patients with Paget's bone disease treated with pamidronate. Bone 32, 15–19 (2003).
Davignon, J. & Leiter, L.A. Ongoing clinical trials of the pleiotropic effects of statins. Vasc. Health Risk Manag. 1, 29–40 (2005).
Rebollo, A., Pou, J. & Alegret, M. Cholesterol lowering and beyond: role of statins in Alzheimer's disease. Aging Health 4, 171–180 (2008).
Docampo, R. & Moreno, S.N. Bisphosphonates as chemotherapeutic agents against trypanosomatid and apicomplexan parasites. Curr. Drug Targets Infect. Disord. 1, 51–61 (2001).
Varela, I. et al. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat. Med. 14, 767–772 (2008).
Wang, X., Hinson, E.R. & Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 (2007).
Stockman, B.J. & Dalvit, C. NMR screening techniques in drug discovery and drug design. Progr. NMR Spectrosc. 41, 187–231 (2002).
Otwinowski, Z. & Minor, W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Kroemer, M., Dreyer, M.K. & Wendt, K.U. APRV—a program for automated data processing, refinement and visualization. Acta Crystallogr. D Biol. Crystallogr. 60, 1679–1682 (2004).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. D54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A A47, 110–119 (1991).
Acknowledgements
We thank G. Hofmann for help with the plasma membrane translocation assay, A. Schmid for help with the scintillation proximity assay, R. Denay for recording the 13C NMR spectra, W. Breitenstein for help in preparing the manuscript and B. Faller for useful discussions. X-ray data collections of the FPPS complexes with 1 to 4, 8 and 10 were performed at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland. We are grateful to the machine and beamline groups whose outstanding efforts have made these experiments possible.
Author information
Authors and Affiliations
Contributions
W.J. initiated the project, led the project team and performed NMR studies together with C.H. J.-M.R. and S.L. performed crystallographic studies. S.C. performed molecular modeling studies. A.M. and X.P. synthesized chemical compounds. M. Geiser, A.S. and R.H. cloned, expressed and purified protein. M. Götte performed the cellular assay, F.B. performed ITC experiments, J.F.G., T.P.R. and S.J.S. performed the biochemical assay. W.J., J.-M.R. and J.R.G. wrote the paper with input from all coauthors.
Corresponding author
Ethics declarations
Competing interests
All authors are past or present full-time employees of Novartis AG and hold stock in the company.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1 & 2 and Supplementary Methods (PDF 479 kb)
Rights and permissions
About this article
Cite this article
Jahnke, W., Rondeau, JM., Cotesta, S. et al. Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery. Nat Chem Biol 6, 660–666 (2010). https://doi.org/10.1038/nchembio.421
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.421
This article is cited by
-
Post-translational modification of KRAS: potential targets for cancer therapy
Acta Pharmacologica Sinica (2021)
-
Human farnesyl pyrophosphate synthase is allosterically inhibited by its own product
Nature Communications (2017)
-
Biophysics in drug discovery: impact, challenges and opportunities
Nature Reviews Drug Discovery (2016)
-
Small-molecule modulation of Ras signaling
Nature Chemical Biology (2014)
-
Ternary complex structures of human farnesyl pyrophosphate synthase bound with a novel inhibitor and secondary ligands provide insights into the molecular details of the enzyme’s active site closure
BMC Structural Biology (2012)