Abstract
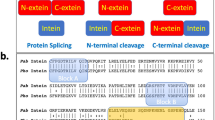
Protein splicing is a post-translational modification in which an intein domain excises itself out of a host protein. Here, we investigate how the steps in the splicing process are coordinated so as to maximize the production of the final splice products and minimize the generation of undesired cleavage products. Our approach has been to prepare a branched intermediate (and analogs thereof) of the Mycobacterium xenopi DNA gyrase A (Mxe GyrA) intein using protein semisynthesis. Kinetic analysis of these molecules indicates that the high fidelity of this protein-splicing reaction results from the penultimate step in the process (intein-succinimide formation) being rate-limiting. NMR experiments indicate that formation of the branched intermediate affects the local structure around the amide bond that is cleaved during succinimide formation. We propose that this structural change reflects a reorganization of the catalytic apparatus to accelerate succinimide formation at the C-terminal splice junction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Noren, C.J., Wang, J. & Perler, F.B. Dissecting the chemistry of protein splicing and its applications. Angew. Chem. Int. Ed. Engl. 39, 450–466 (2000).
Paulus, H. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69, 447–496 (2000).
Perler, F.B. InBase: the intein database. Nucleic Acids Res. 30, 383–384 (2002).
Porter, J.A. et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34 (1996).
Xu, Q., Buckley, D., Guan, C. & Guo, H. Structural insights into the mechanism of intramolecular proteolysis. Cell 98, 651–661 (1999).
Rosenblum, J.S. & Blobel, G. Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. USA 96, 11370–11375 (1999).
Xu, M.-Q. et al. Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation. EMBO J. 13, 5517–5522 (1994).
Xu, M.-Q., Southworth, M.W., Mersha, F.B., Hornstra, L.J. & Perler, F.B. In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell 75, 1371–1377 (1993).
Dawson, P.E., Muir, T.W., Clark-Lewis, I. & Kent, S.B. Synthesis of proteins by native chemical ligation. Science 266, 776–779 (1994).
Poland, B.W., Xu, M.-Q. & Quiocho, F.A. Structural insight into protein splicing mechanism of PI-SceI*. J. Biol. Chem. 275, 16408–16413 (2000).
Ding, Y. et al. Crystal structure of a mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing. J. Biol. Chem. 278, 39133–39142 (2003).
Sun, P. et al. Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal catalytic model without the penultimate histidine and the mechanism of Zn ion inhibition of protein splicing. J. Mol. Biol. 353, 1093–1105 (2005).
Mizutani, R. et al. Protein-splicing reaction via a thiazolidine intermediate: crystal structure of the VMA-1 derived endonuclease bearing the N- and C-terminal propeptides. J. Mol. Biol. 316, 919–929 (2002).
Johnson, M.A. et al. NMR structure of a KlbA intein predursor from Methanococcus jannaschii. Protein Sci. 16, 1316–1328 (2007).
Oeemig, J.S., Aranko, A.S., Djupsjöbacka, J., Heinämäki, K. & Iwaï, H. Solution structure of DnaE intein from Nostoc punctiforme: structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS Lett. 583, 1451–1456 (2009).
Klabunde, T., Sharma, S., Telenti, A., Jacobs, W.R. & Sacchettini, J. Crystal structure of GyrA intein from Mycobacterium xenopi reveal structural basis of protein splicing. Nat. Struct. Biol. 5, 31–36 (1998).
Romanelli, A., Shekhtman, A., Cowburn, D. & Muir, T.W. Semisynthesis of segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at N-extein-intein junction. Proc. Natl. Acad. Sci. USA 101, 6397–6402 (2004).
Xu, M.-Q. & Perler, F.B. The mechanism of protein splicing and its modulation by mutation. EMBO J. 15, 5146–5153 (1996).
Southworth, M.W., Amaya, K., Evans, T.C., Xu, M.Q. & Perler, F.B. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi Gyrase A intein. Biotechniques 27, 110–114 (1999).
Wood, D.W., Wu, W., Belfort, G., Derbyshire, V. & Belfort, M.A. A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 17, 889–892 (1999).
Telenti, A. et al. The Mycobacterium xenopi GyrA protein splicing element: characterization of a minimal intein. J. Bacteriol. 179, 6378–6382 (1997).
Nogami, S., Satow, Y., Ohya, Y. & Anraku, Y. Probing novel elements for protein splicing in the yeast Vma1 protozyme: a study of replacement mutagenesis and intragenic suppression. Genetics 147, 73–85 (1997).
Amitai, G., Callahan, B.P., Stanger, M.J., Belfort, G. & Belfort, M. Modulation of intein activity by its neighboring extein substrates. Proc. Natl. Acad. Sci. USA 106, 11005–11010 (2009).
Martin, D.D., Xu, M.-Q. & Evans, T.C. Characterization of a naturally occurring trans-splicing intein from Synechocystis sp. PCC6803. Biochemistry 40, 1393–1402 (2001).
Mills, K.V., Dorval, D.M. & Lewandowski, K.T. Kinetic analysis of the individual steps of protein splicing for the Pyrococcus abyssi PolII intein. J. Biol. Chem. 280, 2714–2720 (2005).
Zettler, J., Schuetz, V. & Mootz, H.D. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 583, 909–914 (2009).
Muir, T.W., Sondhi, D. & Cole, P.A. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. USA 95, 6705–6710 (1998).
Kawasaki, M., Nogami, S., Satow, Y., Ohya, Y. & Anraku, Y. Identification of three core regions essential for protein splicing of the yeast Vma1 protozyme. A random mutagenesis study of the entire Vma1-derived endonuclease sequence. J. Biol. Chem. 272, 15668–15674 (1997).
Streitwieser, A. & Scannon, P.J. Acidity of hydrocarbons. I. Equilibrium ion pair acidities of thiophene, benzothiophene, thiazole, benzothiazole, and benzofuran toward cesium cyclohexylamine in cyclohexylamine. J. Am. Chem. Soc. 95, 6273–6276 (1973).
Delaglio, F., Torchia, D.A. & Bax, A. Measurment of 15N–13C J coupling in staphylococcal nuclease. J. Biomol. NMR 1, 439–446 (1991).
Cavanagh, J., Fairbrother, W.J., Palmer, A.G. III & Skelton, N.J. Protein NMR Spectroscopy: Principles and Practice (Academic, San Diego, 1996).
Loria, J.P., Berlow, R.B. & Watt, E.D. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc. Chem. Res. 41, 214–221 (2008).
Benkovic, S.J., Hammes, G.G. & Hammes-Schiffer, S. Free-energy landscape of enzyme catalysis. Biochemistry 47, 3317–3321 (2008).
Goodey, N.M. & Benkovic, S.J. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 4, 474–482 (2008).
Harrison, R.K. & Stein, R.L. Mechanistic studies of peptidyl prolyl cys-trans isomerase: evidence for catalysis by distortion. Biochemistry 29, 1684–1689 (1990).
Lopez, X., Mujika, J.I., Blackburn, G.M. & Karplus, M. Alkaline hydrolysis of amide bonds: effect of bond twist and nitrogen pyramidalization. J. Phys. Chem. A 107, 2304–2315 (2003).
Johansson, D.G.A., Macao, B., Sandberg, A. & Härd, T. Protein autoproteolysis: conformational strain linked to the rate of peptide cleavage by the pH dependence of the N→O acyl shift reaction. J. Am. Chem. Soc. 131, 9475–9477 (2009).
Acknowledgements
We thank S.W. Lockless for helpful discussions. This work is supported by US National Institutes of Health grants GM086868 and GM55843 (to T.W.M.) and by the Generalitat de Catalunya (Spain) for the postdoctoral fellowships Beatriu de Pinos (S.F).
Author information
Authors and Affiliations
Contributions
Experiments were designed by S.F. and T.W.M. Semisynthetic constructs and kinetic studies were performed by S.F. NMR experiments were carried out by S.F. and M.G. Preliminary studies with A185C mutant were performed by B.G. Data were analyzed by S.F., M.G., D.C. and T.W.M. The manuscript was prepared by S.F and T.W.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supp Methods and Figures 1–13 (PDF 11707 kb)
Rights and permissions
About this article
Cite this article
Frutos, S., Goger, M., Giovani, B. et al. Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nat Chem Biol 6, 527–533 (2010). https://doi.org/10.1038/nchembio.371
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.371
This article is cited by
-
Nature-inspired protein ligation and its applications
Nature Reviews Chemistry (2023)
-
Intein-mediated protein engineering for biosensor fabrication
BioChip Journal (2016)
-
A Dual-Intein Autoprocessing Domain that Directs Synchronized Protein Co-Expression in Both Prokaryotes and Eukaryotes
Scientific Reports (2015)
-
1H, 13C, and 15N NMR assignments of a Drosophila Hedgehog autoprocessing domain
Biomolecular NMR Assignments (2014)
-
Recent progress in intein research: from mechanism to directed evolution and applications
Cellular and Molecular Life Sciences (2013)