Abstract
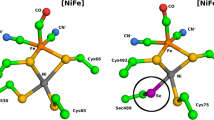
In hydrogenases and many other redox enzymes, the buried active site is connected to the solvent by a molecular channel whose structure may determine the enzyme's selectivity with respect to substrate and inhibitors. The role of these channels has been addressed using crystallography and molecular dynamics, but kinetic data are scarce. Using protein film voltammetry, we determined and then compared the rates of inhibition by CO and O2 in ten NiFe hydrogenase mutants and two FeFe hydrogenases. We found that the rate of inhibition by CO is a good proxy of the rate of diffusion of O2 toward the active site. Modifying amino acids whose side chains point inside the tunnel can slow this rate by orders of magnitude. We quantitatively define the relations between diffusion, the Michaelis constant for H2 and rates of inhibition, and we demonstrate that certain enzymes are slowly inactivated by O2 because access to the active site is slow.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Maynard, E.L. & Lindahl, P.A. Evidence of a molecular tunnel connecting the active sites for CO2 reduction and acetyl-CoA synthesis in acetyl-coa synthase from Clostridium thermoaceticum. J. Am. Chem. Soc. 121, 9221–9222 (1999).
Fontecilla-Camps, J.C., Volbeda, A., Cavazza, C. & Nicolet, Y. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem. Rev. 107, 4273–4303 (2007).
Cohen, J., Kim, K., King, P., Seibert, M. & Schulten, K. Finding gas diffusion pathways in proteins: application to O2 and H2 transport in CpI [FeFe]-hydrogenase and the role of packing defects. Structure 13, 1321–1329 (2005).
Nienhaus, K., Deng, P., Olson, J.S., Warren, J.J. & Nienhaus, G.U. Structural dynamics of myoglobin: ligand migration and binding in valine 68 mutants. J. Biol. Chem. 278, 42532–42544 (2003).
Ruscio, J.Z. et al. Atomic level computational identification of ligand migration pathways between solvent and binding site in myoglobin. Proc. Natl. Acad. Sci. USA 105, 9204–9209 (2008).
Cohen, J. & Schulten, K. O2-migration pathways are not conserved across proteins of a similar fold. Biophys. J. 93, 3591–3600 (2007).
Salomonsson, L., Lee, A., Gennis, R.B. & Brzezinski, P. A single-amino-acid lid renders a gas-tight compartment within a membrane-bound transporter. Proc. Natl. Acad. Sci. USA 101, 11617–11621 (2004).
Leroux, F. et al. Experimental approaches to kinetics of gas diffusion in hydrogenase. Proc. Natl. Acad. Sci. USA 105, 11188–11193 (2008).
Vincent, K.A. et al. Electrocatalytic hydrogen oxidation by an enzyme at high carbon monoxide or oxygen levels. Proc. Natl. Acad. Sci. USA 102, 16951–16954 (2005).
Vincent, K.A. et al. Enzymatic catalysis on conducting graphite particles. Nat. Chem. Biol. 3, 761–762 (2007).
Karyakin, A.A. et al. The limiting performance characteristics in bioelectrocatalysis of hydrogenase enzymes. Angew. Chem. Int. Ed. 46, 7244–7246 (2007).
Hambourger, M. et al. [FeFe]-hydrogenase-catalyzed H2 production in a photo-electrochemical biofuel cell. J. Am. Chem. Soc. 130, 2015–2022 (2008).
Ghirardi, M.L., Dubini, A., Yu, J. & Maness, P.-C. Photobiological hydrogen-producing systems. Chem. Soc. Rev. 38, 52–61 (2009).
Volbeda, A., Montet, Y., Vernède, X., Hatchikian, E.C. & Fontecilla-Camps, J.C. High-resolution crystallographic analysis of Desulfovibrio fructosovorans NiFe hydrogenase. Int. J. Hydrogen Energy 27, 1449–1461 (2002).
Buhrke, T., Lenz, O., Krauss, N. & Friedrich, B. Oxygen tolerance of the H2-sensing [NiFe] hydrogenase from Ralstonia eutropha H16 is based on limited access of oxygen to the active site. J. Biol. Chem. 280, 23791–23796 (2005).
Duché, O., Elsen, S., Cournac, L. & Colbeau, A. Enlarging the gas access channel to the active site renders the regulatory hydrogenase HupUV of Rhodobacter capsulatus O2 sensitive without affecting its transductory activity. FEBS J. 272, 3899–3908 (2005).
Ludwig, M., Cracknell, J.A., Vincent, K.A., Armstrong, F.A. & Lenz, O. Oxygen-tolerant H2 oxidation by membrane-bound [NiFe]-hydrogenases of Ralstonia species: coping with low-level H2 in air. J. Biol. Chem. 284, 465–477 (2009).
Dementin, S. et al. Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant. J. Am. Chem. Soc. 131, 10156–10164 (2009).
Baffert, C. et al. Hydrogen-activating enzymes: activity does not correlate with oxygen-sensitivity. Angew. Chem. Int. Ed. 47, 2052–2055 (2008).
Vincent, K.A., Parkin, A. & Armstrong, F.A. Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem. Rev. 107, 4366–4413 (2007).
Léger, C., Dementin, S., Bertrand, P., Rousset, M. & Guigliarelli, B. Inhibition and aerobic inactivation kinetics of Desulfovibrio fructosovorans NiFe hydrogenases studied by protein film voltammetry. J. Am. Chem. Soc. 126, 12162–12172 (2004).
Léger, C. & Bertrand, P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem. Rev. 108, 2379–2438 (2008).
Guigliarelli, B. et al. Structural organization of the Ni and [4Fe-4S] centers in the active form of Desulfovibrio gigas hydrogenase. Analysis of the magnetic interactions by electron paramagnetic resonance spectroscopy. Biochemistry 34, 4781–4790 (1995).
Almeida, M.G. et al. A needle in a haystack: the active site of the membrane-bound complex cytochrome c nitrite reductase. FEBS Lett. 581, 284–288 (2007).
Rohlfs, R.J., Olson, J.S. & Gibson, Q.H. A comparison of the geminate recombination kinetics of several monomeric heme proteins. J. Biol. Chem. 263, 1803–1813 (1988).
Riistama, S., Puustinen, A., Verkhovsky, M.I., Morgan, J.E. & Wikstrom, M. Binding of O2 and its reduction are both retarded by replacement of valine 279 by isoleucine in cytochrome c oxidase from Paracoccus denitrificans. Biochemistry 39, 6365–6372 (2000).
Chen, L., Lyubimov, A.Y., Brammer, L., Vrielink, A. & Sampson, N.S. The binding and release of oxygen and hydrogen peroxide are directed by a hydrophobic tunnel in cholesterol oxidase. Biochemistry 47, 5368–5377 (2008).
Barney, B.M., Yurth, M.G., Dos Santos, P.C., Dean, D.R. & Seefeldt, L.C. A substrate channel in the nitrogenase MoFe protein. J. Biol. Inorg. Chem. 14, 1015–1022 (2009).
Dementin, S. et al. A glutamate is the essential proton transfer gate during the catalytic cycle of the NiFe hydrogenase. J. Biol. Chem. 279, 10508–10513 (2004).
Dementin, S. et al. Changing the ligation of the distal [4Fe4S] cluster in NiFe hydrogenase impairs inter- and intramolecular electron transfers. J. Am. Chem. Soc. 128, 5209–5218 (2006).
Rousset, M. New shuttle vectors for the introduction of cloned DNA in Desulfovibrio. Plasmid 39, 114–122 (1998).
Hatchikian, E.C., Forget, N., Fernandez, V.M., Williams, R. & Cammack, R. Further characterization of the [Fe]-hydrogenase from Desulfovibrio desulfuricans ATCC 7757. Eur. J. Biochem. 209, 357–365 (1992).
Fourmond, V. et al. Correcting for electrocatalyst desorption and inactivation in chronoamperometry experiments. Anal. Chem. 81, 2962–2968 (2009).
Rousset, M. et al. [3Fe-4S] to [4Fe-4S] cluster conversion in Desulfovibrio fructosovorans [NiFe] hydrogenase by site-directed mutagenesis. Proc. Natl. Acad. Sci. USA 95, 11625–11630 (1998).
Fourmond, V. et al. SOAS: a free software to analyse electrochemical data and other one-dimensional signals. Bioelectrochem. 76, 141–147 (2009).
Demuez, M. et al. Complete activity profile of Clostridium acetobutylicum [FeFe]-hydrogenase and kinetic parameters for endogenous redox partners. FEMS Microbiol. Lett. 275, 113–121 (2007).
Acknowledgements
This work was funded by the Centre National de la Recherche Scientifique, Commissariat à l'Energie Atomique, Agence Nationale de la Recherche, the University of Provence and the City of Marseilles, and supported by the Pôle de Compétitivité Capénergies. The Groupe de Recherche 2977 (“Bio-hydrogène”) paid the publication fees for this article.
Author information
Authors and Affiliations
Contributions
P.-P.L. designed mutants of the NiFe enzyme, performed mutagenesis, carried out protein purification, solution assays and electrochemical measurements, and analyzed data, with the support of M.R. and C.L. F.L. performed electrochemical measurements on several forms of the NiFe hydrogenase (WT, L122M V74M, V74M, L122F V74I). B.B. characterized by EPR the NiFe hydrogenase mutants, with the support of B.G. S.D. designed mutants of the NiFe enzyme, performed mutagenesis, carried out protein purification and solution assays, and interpreted studies, with the support of M.R. C.B. performed the electrochemical characterization of the two FeFe hydrogenases and analyzed the data. T.L. purified the FeFe hydrogenase from C. acetobutylicum and assayed its activity, with the support and advice of I.M.-S. and P.S. V.F. contributed to modeling. P.C. characterized the V74W mutant, and analyzed the data. C.C. purified the FeFe hydrogenase from D. desulfuricans, with the support of J.C.F.-C. P.-P.L., S.D., B.B., C.B., M.R., B.G., P.B. and C.L. co-designed research. S.D., P.B. and C.L. conceptualized, analyzed and interpreted all studies and co-wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 183 kb)
Rights and permissions
About this article
Cite this article
Liebgott, PP., Leroux, F., Burlat, B. et al. Relating diffusion along the substrate tunnel and oxygen sensitivity in hydrogenase. Nat Chem Biol 6, 63–70 (2010). https://doi.org/10.1038/nchembio.276
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.276
This article is cited by
-
Protein film electrochemistry
Nature Reviews Methods Primers (2023)
-
O2-tolerant CO dehydrogenase via tunnel redesign for the removal of CO from industrial flue gas
Nature Catalysis (2022)
-
Microbial oxidation of atmospheric trace gases
Nature Reviews Microbiology (2022)
-
A safety cap protects hydrogenase from oxygen attack
Nature Communications (2021)
-
Isolation of an archaeon at the prokaryote–eukaryote interface
Nature (2020)