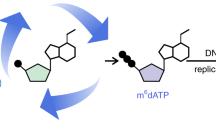
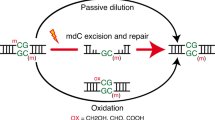
Abstract
Tet enzymes oxidize 5-methyl-deoxycytidine (mdC) to 5-hydroxymethyl-dC (hmdC), 5-formyl-dC (fdC) and 5-carboxy-dC (cadC) in DNA. It was proposed that fdC and cadC deformylate and decarboxylate, respectively, to dC over the course of an active demethylation process. This would re-install canonical dC bases at previously methylated sites. However, whether such direct C–C bond cleavage reactions at fdC and cadC occur in vivo remains an unanswered question. Here we report the incorporation of synthetic isotope- and (R)-2′-fluorine-labeled dC and fdC derivatives into the genome of cultured mammalian cells. Following the fate of these probe molecules using UHPLC–MS/MS provided quantitative data about the formed reaction products. The data show that the labeled fdC probe is efficiently converted into the corresponding labeled dC, most likely after its incorporation into the genome. Therefore, we conclude that fdC undergoes C–C bond cleavage in stem cells, leading to the direct re-installation of unmodified dC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Smith, Z.D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet 14, 204–220 (2013).
Schübeler, D. Function and information content of DNA methylation. Nature 517, 321–326 (2015).
Jeltsch, A. & Jurkowska, R.Z. Allosteric control of mammalian DNA methyltransferases – a new regulatory paradigm Nucleic Acids Res. 44, 8556–8575 (2016).
Tahiliani, M. et al. Conversion of 5-methylcytosine to -hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Pfaffeneder, T. et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed. Engl. 50, 7008–7012 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Globisch, D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5, e15367 (2010).
Münzel, M., Globisch, D. & Carell, T. 5-Hydroxymethylcytosine, the sixth base of the genome. Angew. Chem. Int. Ed. Engl. 50, 6460–6468 (2011).
Pfaffeneder, T. et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 10, 574–581 (2014).
Wagner, M. et al. Age-dependent levels of 5-methyl-, 5-hydroxymethyl-, and 5-formylcytosine in human and mouse brain tissues. Angew. Chem. Int. Edn Engl. 54, 12511–12514 (2015).
Branco, M.R., Ficz, G. & Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 (2011).
Wu, H. & Zhang, Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25, 2436–2452 (2011).
Bachman, M. et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 11, 555–557 (2015).
Su, M. et al. 5-Formylcytosine could be a semipermanent base in specific genome sites. Angew. Chem. Int. Ed. Engl. 55, 11797–11800 (2016).
Raiber, E.A. et al. 5-Formylcytosine alters the structure of the DNA double helix. Nat. Struct. Mol. Biol. 22, 44–49 (2015).
Song, C.X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013).
Kellinger, M.W. et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 19, 831–833 (2012).
Zhu, C. et al. Single-cell 5-formylcytosine landscapes of mammalian early embryos and ESCs at single-base resolution. Cell Stem Cell 20, 720–731.e5 (2017).
Hill, P.W., Amouroux, R. & Hajkova, P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics 104, 324–333 (2014).
Wu, X., Inoue, A., Suzuki, T. & Zhang, Y. Simultaneous mapping of active DNA demethylation and sister chromatid exchange in single cells. Genes Dev. 31, 511–523 (2017).
Maiti, A. & Drohat, A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 (2011).
Guo, F. et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459 (2014).
Wu, S.C. & Zhang, Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620 (2010).
Schiesser, S. et al. Deamination, oxidation, and C-C bond cleavage reactivity of 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine. J. Am. Chem. Soc. 135, 14593–14599 (2013).
Liutkevičiūtė, Z. et al. Direct decarboxylation of 5-carboxylcytosine by DNA C5-methyltransferases. J. Am. Chem. Soc. 136, 5884–5887 (2014).
Schiesser, S. et al. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew. Chem. Int. Ed. Engl. 51, 6516–6520 (2012).
Jekunen, A. & Vilpo, J.A. 5-Methyl-2′-deoxycytidine. Metabolism and effects on cell lethality studied with human leukemic cells in vitro. Mol. Pharmacol. 25, 431–435 (1984).
Vilpo, J.A. & Vilpo, L.M. Biochemical mechanisms by which reutilization of DNA 5-methylcytosine is prevented in human cells. Mutat. Res. 256, 29–35 (1991).
Schröder, A.S. et al. Synthesis of (R)-configured 2′-fluorinated mC, hmC, fC, and caC phosphoramidites and oligonucleotides. Org. Lett. 18, 4368–4371 (2016).
Schröder, A.S. et al. 2′-(R)-Fluorinated mC, hmC, fC and caC triphosphates are substrates for DNA polymerases and TET-enzymes. Chem. Commun. (Camb.) 52, 14361–14364 (2016).
Blaschke, K. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 (2013).
Minor, E.A., Court, B.L., Young, J.I. & Wang, G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 288, 13669–13674 (2013).
Yin, R. et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135, 10396–10403 (2013).
Wu, X. & Zhang, Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534 (2017).
Hargrove, T.Y. et al. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14alpha-demethylase (CYP51) from Leishmania infantum. J. Biol. Chem. 286, 26838–26848 (2011).
Lepesheva, G.I. et al. CYP51: A major drug target in the cytochrome P450 superfamily. Lipids 43, 1117–1125 (2008).
Aukema, K.G. et al. Cyanobacterial aldehyde deformylase oxygenation of aldehydes yields n-1 aldehydes and alcohols in addition to alkanes. ACS Catal. 3, 2228–2238 (2013).
Jia, C. et al. Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases. Protein Cell 6, 55–67 (2015).
Fujihashi, M., Mnpotra, J.S., Mishra, R.K., Pai, E.F. & Kotra, L.P. Orotidine monophosphate decarboxylase--a fascinating workhorse enzyme with therapeutic potential. J. Genet. Genomics 42, 221–234 (2015).
Smiley, J.A., Angelot, J.M., Cannon, R.C., Marshall, E.M. & Asch, D.K. Radioactivity-based and spectrophotometric assays for isoorotate decarboxylase: identification of the thymidine salvage pathway in lower eukaryotes. Anal. Biochem. 266, 85–92 (1999).
Xu, S. et al. Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase. Cell Res. 23, 1296–1309 (2013).
Kim, H. et al. Modulation of β-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat. Commun. 4, 2403 (2013).
Toyooka, Y., Shimosato, D., Murakami, K., Takahashi, K. & Niwa, H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909–918 (2008).
Shirane, K. et al. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev. Cell 39, 87–103 (2016).
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S. & Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532 (2011).
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006).
Hu, X. et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14, 512–522 (2014).
Dawlaty, M.M. et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29, 102–111 (2014).
Liu, N. et al. Intrinsic and extrinsic connections of Tet3 dioxygenase with CXXC zinc finger modules. PLoS One 8, e62755 (2013).
Cao, H. & Wang, Y. Collisionally activated dissociation of protonated 2′-deoxycytidine, 2′-deoxyuridine, and their oxidatively damaged derivatives. J. Am. Soc. Mass Spectrom. 17, 1335–1341 (2006).
Spruijt, C.G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013).
Wang, J. et al. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal. Chem. 83, 2201–2209 (2011).
Dietmair, S., Timmins, N.E., Gray, P.P., Nielsen, L.K. & Krömer, J.O. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal. Biochem. 404, 155–164 (2010).
Acknowledgements
Tet TKO mESC lines were kindly provided by G.-L. Xu (Shanghai Institutes for Biological Sciences) and R. Jaenisch (Whitehead Institute, MIT, Cambridge). We are grateful to M. Okano and H. Niwa (both at Kumamoto University, Japan) for providing the Dnmt TKO mESC line and the Oct4-YFP reporter cell line, respectively. A.S.S. is supported by a fellowship from the Fonds der Chemischen Industrie. We thank the Deutsche Forschungsgemeinschaft for financial support through the programs: SFB749 (TP A4), SFB1032 (TP A5), SPP1784 and CA275-11/1. We thank the European Union Horizon 2020 program for funding the ERC Advanced project EPiR (741912). Further support is acknowledged from the Excellence Cluster CiPSM (Center for Integrated Protein Science).
Author information
Authors and Affiliations
Contributions
K.I. developed and performed the UHPLC–MS/MS studies. R.R. and A.S.S. synthesized the fluorinated and isotopically labeled nucleosides. A.K. designed and performed cell culture work. F.S. designed, supervised and performed cell culture work. O.K. and J.S. analyzed feeding studies of isotopically labeled dC. S.F. contributed to experiments for the analysis of soluble nucleoside pools. M.M. supervised the biochemical work, interpreted and discussed results. T.C. designed and supervised the study. All members discussed results, interpreted data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–4, Supplementary Figures 1–13 (PDF 1111 kb)
Supplementary Note
Synthetic Procedures (PDF 1589 kb)
Rights and permissions
About this article
Cite this article
Iwan, K., Rahimoff, R., Kirchner, A. et al. 5-Formylcytosine to cytosine conversion by C–C bond cleavage in vivo. Nat Chem Biol 14, 72–78 (2018). https://doi.org/10.1038/nchembio.2531
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2531
This article is cited by
-
Redirected nuclear glutamate dehydrogenase supplies Tet3 with α-ketoglutarate in neurons
Nature Communications (2021)
-
Reversal of nucleobase methylation by dioxygenases
Nature Chemical Biology (2020)
-
Active turnover of genomic methylcytosine in pluripotent cells
Nature Chemical Biology (2020)
-
Isotope-dilution mass spectrometry for exact quantification of noncanonical DNA nucleosides
Nature Protocols (2019)
-
Methods for detection of cytosine and thymine modifications in DNA
Nature Reviews Chemistry (2018)