Abstract
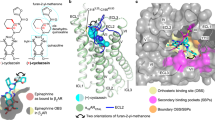
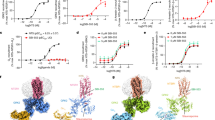
Development of biased ligands targeting G protein-coupled receptors (GPCRs) is a promising approach for current drug discovery. Although structure-based drug design of biased agonists remains challenging even with an abundance of GPCR crystal structures, we present an approach for translating GPCR structural data into β-arrestin-biased ligands for aminergic GPCRs. We identified specific amino acid–ligand contacts at transmembrane helix 5 (TM5) and extracellular loop 2 (EL2) responsible for Gi/o and β-arrestin signaling, respectively, and targeted those residues to develop biased ligands. For these ligands, we found that bias is conserved at other aminergic GPCRs that retain similar residues at TM5 and EL2. Our approach provides a template for generating arrestin-biased ligands by modifying predicted ligand interactions that block TM5 interactions and promote EL2 interactions. This strategy may facilitate the structure-guided design of arrestin-biased ligands at other GPCRs, including polypharmacological biased ligands.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Overington, J.P., Al-Lazikani, B. & Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Rask-Andersen, M., Almén, M.S. & Schiöth, H.B. Trends in the exploitation of novel drug targets. Nat. Rev. Drug Discov. 10, 579–590 (2011).
Wacker, D., Stevens, R.C. & Roth, B.L. How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017).
Urban, J.D. et al. Functional selectivity and classical concepts of quantitative pharmacology. J. Pharmacol. Exp. Ther. 320, 1–13 (2007).
DeWire, S.M., Ahn, S., Lefkowitz, R.J. & Shenoy, S.K. β-arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510 (2007).
Shukla, A.K., Xiao, K. & Lefkowitz, R.J. Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem. Sci. 36, 457–469 (2011).
Violin, J.D., Crombie, A.L., Soergel, D.G. & Lark, M.W. Biased ligands at G-protein-coupled receptors: promise and progress. Trends Pharmacol. Sci. 35, 308–316 (2014).
Allen, J.A. et al. Discovery of β-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc. Natl. Acad. Sci. USA 108, 18488–18493 (2011).
Soergel, D.G. et al. Biased agonism of the μ-opioid receptor by TRV130 increases analgesia and reduces on-target adverse effects versus morphine: A randomized, double-blind, placebo-controlled, crossover study in healthy volunteers. Pain 155, 1829–1835 (2014).
Violin, J.D. et al. Selectively engaging β-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J. Pharmacol. Exp. Ther. 335, 572–579 (2010).
Urs, N.M. et al. Targeting β-arrestin2 in the treatment of L-DOPA-induced dyskinesia in Parkinson's disease. Proc. Natl. Acad. Sci. USA 112, E2517–E2526 (2015).
Charfi, I., Audet, N., Bagheri Tudashki, H. & Pineyro, G. Identifying ligand-specific signalling within biased responses: focus on δ opioid receptor ligands. Br. J. Pharmacol. 172, 435–448 (2015).
Manglik, A. et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature 537, 185–190 (2016).
DeWire, S.M. et al. A G protein-biased ligand at the μ-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphine. J. Pharmacol. Exp. Ther. 344, 708–717 (2013).
Gesty-Palmer, D. et al. β-arrestin-selective G protein-coupled receptor agonists engender unique biological efficacy in vivo. Mol. Endocrinol. 27, 296–314 (2013).
Masri, B. et al. Antagonism of dopamine D2 receptor/β-arrestin 2 interaction is a common property of clinically effective antipsychotics. Proc. Natl. Acad. Sci. USA 105, 13656–13661 (2008).
Tchernychev, B. et al. Discovery of a CXCR4 agonist pepducin that mobilizes bone marrow hematopoietic cells. Proc. Natl. Acad. Sci. USA 107, 22255–22259 (2010).
Cherezov, V. et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007).
Chien, E.Y. et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330, 1091–1095 (2010).
Rasmussen, S.G. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Ring, A.M. et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502, 575–579 (2013).
Wacker, D. et al. Structural features for functional selectivity at serotonin receptors. Science 340, 615–619 (2013).
Wang, C. et al. Structural basis for molecular recognition at serotonin receptors. Science 340, 610–614 (2013).
Kapur, S. & Remington, G. Atypical antipsychotics: new directions and new challenges in the treatment of schizophrenia. Annu. Rev. Med. 52, 503–517 (2001).
Wadenberg, M.L., Soliman, A., VanderSpek, S.C. & Kapur, S. Dopamine D(2) receptor occupancy is a common mechanism underlying animal models of antipsychotics and their clinical effects. Neuropsychopharmacology 25, 633–641 (2001).
Shapiro, D.A. et al. Aripiprazole, a novel atypical antipsychotic drug with a unique and robust pharmacology. Neuropsychopharmacology 28, 1400–1411 (2003).
Chen, X. et al. Discovery of G protein-biased D2 dopamine receptor partial agonists. J. Med. Chem. 59, 10601–10618 (2016).
Park, S.M. et al. Effects of β-arrestin-biased dopamine D2 receptor ligands on schizophrenia-like behavior in hypoglutamatergic mice. Neuropsychopharmacology 41, 704–715 (2016).
Ambrosio, C., Molinari, P., Cotecchia, S. & Costa, T. Catechol-binding serines of β2-adrenergic receptors control the equilibrium between active and inactive receptor states. Mol. Pharmacol. 57, 198–210 (2000).
Isogai, S. et al. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530, 237–241 (2016).
Warne, T. et al. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature 469, 241–244 (2011).
Neve, K.A. & Wiens, B.L. Four ways of being an agonist: multiple sequence determinants of efficacy at D2 dopamine receptors. Biochem. Soc. Trans. 23, 112–116 (1995).
Wiens, B.L., Nelson, C.S. & Neve, K.A. Contribution of serine residues to constitutive and agonist-induced signaling via the D2S dopamine receptor: evidence for multiple, agonist-specific active conformations. Mol. Pharmacol. 54, 435–444 (1998).
Fowler, J.C., Bhattacharya, S., Urban, J.D., Vaidehi, N. & Mailman, R.B. Receptor conformations involved in dopamine D(2L) receptor functional selectivity induced by selected transmembrane-5 serine mutations. Mol. Pharmacol. 81, 820–831 (2012).
Wacker, D. et al. Crystal structure of an LSD-bound human serotonin receptor. Cell 168, 377–389.e12 (2017).
Chen, X. et al. Structure-functional selectivity relationship studies of β-arrestin-biased dopamine D2 receptor agonists. J. Med. Chem. 55, 7141–7153 (2012).
Christopher, J.A. et al. Biophysical fragment screening of the β1-adrenergic receptor: identification of high affinity arylpiperazine leads using structure-based drug design. J. Med. Chem. 56, 3446–3455 (2013).
Kling, R.C., Tschammer, N., Lanig, H., Clark, T. & Gmeiner, P. Active-state model of a dopamine D2 receptor-Gαi complex stabilized by aripiprazole-type partial agonists. PLoS One 9, e100069 (2014).
Luedtke, R.R. et al. Comparison of the binding and functional properties of two structurally different D2 dopamine receptor subtype selective compounds. ACS Chem. Neurosci. 3, 1050–1062 (2012).
Kroeze, W.K. et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015).
Klein Herenbrink, C. et al. The role of kinetic context in apparent biased agonism at GPCRs. Nat. Commun. 7, 10842 (2016).
Ahuja, S. et al. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat. Struct. Mol. Biol. 16, 168–175 (2009).
Kuwayama, S., Imai, H., Hirano, T., Terakita, A. & Shichida, Y. Conserved proline residue at position 189 in cone visual pigments as a determinant of molecular properties different from rhodopsins. Biochemistry 41, 15245–15252 (2002).
Warne, T. & Tate, C.G. The importance of interactions with helix 5 in determining the efficacy of β-adrenoceptor ligands. Biochem. Soc. Trans. 41, 159–165 (2013).
Deupi, X. & Standfuss, J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 21, 541–551 (2011).
Roth, B.L., Sheffler, D.J. & Kroeze, W.K. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 (2004).
Besnard, J. et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012).
Huang, W. et al. Structural insights into μ-opioid receptor activation. Nature 524, 315–321 (2015).
Kobilka, B. & Schertler, G.F. New G-protein-coupled receptor crystal structures: insights and limitations. Trends Pharmacol. Sci. 29, 79–83 (2008).
Motulsky, H.J. & Mahan, L.C. The kinetics of competitive radioligand binding predicted by the law of mass action. Mol. Pharmacol. 25, 1–9 (1984).
Kenakin, T., et al. A simple method for quantifying functional selectivity and agonist bias. ACS Chem. Neurosci. 3, 193–203 (2012).
Weiss, D.R. et al. Conformation guides molecular efficacy in docking screens of activated β-2 adrenergic G protein coupled receptor. ACS Chem. Biol. 8, 1018–1026 (2013).
Coleman, R.G., Sterling, T. & Weiss, D.R. SAMPL4 & DOCK3.7: lessons for automated docking procedures. J. Comput. Aided Mol. Des. 28, 201–209 (2014).
Lomize, M.A., Lomize, A.L., Pogozheva, I.D. & Mosberg, H.I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006).
Zhang, L. & Hermans, J. Hydrophilicity of cavities in proteins. Proteins 24, 433–438 (1996).
Best, R.B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Vanommeslaeghe, K. & MacKerell, A.D. Jr. Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012).
Betz, R.M. & Walker, R.C. Paramfit: automated optimization of force field parameters for molecular dynamics simulations. J. Comput. Chem. 36, 79–87 (2015).
Salomon-Ferrer, R., Götz, A.W., Poole, D., Le Grand, S. & Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Roe, D.R. & Cheatham, T.E. III. PTRAJ and CPPTRAJ: software for processing and analysis of molecular synamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Acknowledgements
We thank S. Hollingsworth for assistance with simulation analysis and A.J. Venkatakrishnan for assistance with simulation setup. We thank R. Axel at Columbia University for the HTLA cells and M. Bouvier at Université de Montréal for BRET constructs. This work was supported by the National Institutes of Health (NIH) grant U19MH082441 (to B.L.R. and J.J.), R01MH112205 (to B.L.R.), R01NS100930 (to J.J.), the National Institute of Mental Health Psychoactive Drug Screening Program (NIMH PDSP; to B.L.R.), the Michael Hooker Chair for Protein Therapeutics and Translational Proteomics (to B.L.R.), the American Cancer Society postdoctoral fellowship PF-14-021-01-CDD (to K.V.B.), by NIH grant GM59957 (to B.K.S.), by Pfizer, Inc. (R.O.D.), by a Terman Faculty Fellowship (to R.O.D.), and by a National Science Foundation Graduate Research Fellowship (to R.M.B.).
Author information
Authors and Affiliations
Contributions
J.D.M. designed experiments, performed mutagenesis, ligand-binding and signaling studies, analyzed the data, and wrote the manuscript. K.V.B. designed and synthesized all ligands, performed analytical chemical analysis and wrote the manuscript. B.K. performed and analyzed MD simulations, used the results to design ligands, and wrote the manuscript. K.R. assisted with mutagenesis and signaling studies. B.K., J.K., and B.L.K. built the D2 homology model. J.K. performed the docking experiments and edited the manuscript. R.M.B. determined ligand parameters and performed preliminary MD simulations. B.K.S. supervised the docking experiments and edited the manuscript. R.O.D. supervised the MD simulation studies and helped prepare the manuscript. J.J. supervised ligand synthesis, designed experiments and edited the manuscript. B.L.R. designed the experiments, was responsible for the overall project strategy and management and prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–6 and Supplementary Figures 1–10 (PDF 1763 kb)
Supplementary Note 1
Synthetic chemistry procedures (PDF 504 kb)
Rights and permissions
About this article
Cite this article
McCorvy, J., Butler, K., Kelly, B. et al. Structure-inspired design of β-arrestin-biased ligands for aminergic GPCRs. Nat Chem Biol 14, 126–134 (2018). https://doi.org/10.1038/nchembio.2527
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2527
This article is cited by
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Molecular basis of human trace amine-associated receptor 1 activation
Nature Communications (2024)
-
Constrained catecholamines gain β2AR selectivity through allosteric effects on pocket dynamics
Nature Communications (2023)
-
Mechanism of activation and biased signaling in complement receptor C5aR1
Cell Research (2023)
-
Design and structural validation of peptide–drug conjugate ligands of the kappa-opioid receptor
Nature Communications (2023)