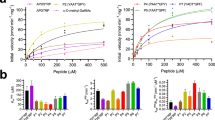
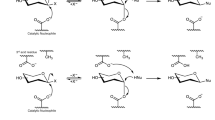
Abstract
O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) is an essential human glycosyltransferase that adds O-GlcNAc modifications to numerous proteins. However, little is known about the mechanism with which OGT recognizes various protein substrates. Here we report on GlcNAc electrophilic probes (GEPs) to expedite the characterization of OGT–substrate recognition. Data from mass spectrometry, X-ray crystallization, and biochemical and radiolabeled kinetic assays support the application of GEPs to rapidly report the impacts of OGT mutations on protein substrate or sugar binding and to discover OGT residues crucial for protein recognition. Interestingly, we found that the same residues on the inner surface of the N-terminal domain contribute to OGT interactions with different protein substrates. By tuning reaction conditions, a GEP enables crosslinking of OGT with acceptor substrates in situ, affording a unique method to discover genuine substrates that weakly or transiently interact with OGT. Hence, GEPs provide new strategies to dissect OGT–substrate binding and recognition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Vocadlo, D.J. O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr. Opin. Chem. Biol. 16, 488–497 (2012).
Hardivillé, S. & Hart, G.W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20, 208–213 (2014).
Hart, G.W., Slawson, C., Ramirez-Correa, G. & Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 (2011).
Capotosti, F. et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 (2011).
Ma, Z., Vocadlo, D.J. & Vosseller, K. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J. Biol. Chem. 288, 15121–15130 (2013).
Shafi, R. et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97, 5735–5739 (2000).
Howerton, C.L. & Bale, T.L. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 111, 9639–9644 (2014).
Ma, J. & Hart, G.W. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteomics 10, 365–380 (2013).
de Queiroz, R.M., Carvalho, E. & Dias, W.B. O-GlcNAcylation: the sweet side of the cancer. Front. Oncol. 4, 132 (2014).
Yuzwa, S.A. & Vocadlo, D.J. O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer's disease and beyond. Chem. Soc. Rev. 43, 6839–6858 (2014).
Laczy, B. et al. Protein O-GlcNAcylation: a new signaling paradigm for the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 296, H13–H28 (2009).
Ma, J. & Hart, G.W. O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics 11, 8 (2014).
Levine, Z.G. & Walker, S. The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells? Annu. Rev. Biochem. 85, 631–657 (2016).
Iyer, S.P.N. & Hart, G.W. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 278, 24608–24616 (2003).
Lazarus, M.B. et al. Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol. 8, 966–968 (2012).
Lazarus, M.B., Nam, Y., Jiang, J., Sliz, P. & Walker, S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 (2011).
Pierce, M.M., Raman, C.S. & Nall, B.T. Isothermal titration calorimetry of protein-protein interactions. Methods 19, 213–221 (1999).
Hoa, X.D., Kirk, A.G. & Tabrizian, M. Towards integrated and sensitive surface plasmon resonance biosensors: a review of recent progress. Biosens. Bioelectron. 23, 151–160 (2007).
Leitner, A., Faini, M., Stengel, F. & Aebersold, R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 (2016).
Jiang, J., Lazarus, M.B., Pasquina, L., Sliz, P. & Walker, S. A neutral diphosphate mimic crosslinks the active site of human O-GlcNAc transferase. Nat. Chem. Biol. 8, 72–77 (2011).
Lairson, L.L., Henrissat, B., Davies, G.J. & Withers, S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008).
Kaur, P., Kiselar, J., Yang, S. & Chance, M.R. Quantitative protein topography analysis and high-resolution structure prediction using hydroxyl radical labeling and tandem-ion mass spectrometry (MS). Mol. Cell. Proteomics 14, 1159–1168 (2015).
Pathak, S. et al. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 22, 744–750 (2015).
Kreppel, L.K. & Hart, G.W. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 274, 32015–32022 (1999).
Mayer, A., Gloster, T.M., Chou, W.K., Vocadlo, D.J. & Tanner, M.E. 6″-Azido-6″-deoxy-UDP-N-acetylglucosamine as a glycosyltransferase substrate. Bioorg. Med. Chem. Lett. 21, 1199–1201 (2011).
Chuh, K.N., Zaro, B.W., Piller, F., Piller, V. & Pratt, M.R. Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J. Am. Chem. Soc. 136, 12283–12295 (2014).
Davis, L.I. & Blobel, G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84, 7552–7556 (1987).
Schimpl, M. et al. O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat. Chem. Biol. 8, 969–974 (2012).
Kumari, M. et al. Exploring reaction pathways for O-GlcNAc transferase catalysis. A string method study. J. Phys. Chem. B 119, 4371–4381 (2015).
Lazarus, B.D., Love, D.C. & Hanover, J.A. Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16, 415–421 (2006).
Rexach, J.E. et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat. Chem. Biol. 6, 645–651 (2010).
Khidekel, N. et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 (2007).
Zhao, P. et al. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J. Proteome Res. 10, 4088–4104 (2011).
Lee, A. et al. Combined antibody/lectin enrichment identifies extensive changes in the O-GlcNAc sub-proteome upon oxidative stress. J. Proteome Res. 15, 4318–4336 (2016).
Hahne, H. et al. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J. Proteome Res. 12, 927–936 (2013).
Hahne, H., Gholami, A.M. & Kuster, B. Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol. Cell. Proteomics 11, 843–850 (2012).
Kamemura, K., Hayes, B.K., Comer, F.I. & Hart, G.W. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: alternative glycosylation/phosphorylation of THR-58, a known mutational hot spot of c-Myc in lymphomas, is regulated by mitogens. J. Biol. Chem. 277, 19229–19235 (2002).
Malaker, S.A. et al. Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol. Res. 5, 376–384 (2017).
Phueaouan, T. et al. Aberrant O-GlcNAc-modified proteins expressed in primary colorectal cancer. Oncol. Rep. 30, 2929–2936 (2013).
Lazarus, M.B. et al. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342, 1235–1239 (2013).
Wells, L. et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791–804 (2002).
Vosseller, K. et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 (2006).
Vocadlo, D.J., Hang, H.C., Kim, E.-J., Hanover, J.A. & Bertozzi, C.R. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. USA 100, 9116–9121 (2003).
Sakaidani, Y. et al. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2, 583 (2011).
Müller, R., Jenny, A. & Stanley, P. The EGF repeat-specific O-GlcNAc-transferase Eogt interacts with notch signaling and pyrimidine metabolism pathways in Drosophila. PLoS One 8, e62835 (2013).
Xiao, H. & Wu, R. Global and site-specific analysis revealing unexpected and extensive protein S-GlcNAcylation in human cells. Anal. Chem. 89, 3656–3663 (2017).
Zaro, B.W., Yang, Y.Y., Hang, H.C. & Pratt, M.R. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc. Natl. Acad. Sci. USA 108, 8146–8151 (2011).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Boersema, P.J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A.J.R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
UniProt Consortium. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42, D191–D198 (2014).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We thank S. Walker's lab at Harvard Medical School for kindly sharing the expression plasmids of OGT, OGT4.5, OGA, and NUP62. We would like to acknowledge the research funding support from University of Wisconsin–Madison (to J.J.), a Vilas Research Investigator Award (to J.J.), NIH R01 GM121718 (to J.J.), NIH R21 AG055377 (to L.L.), NIH R01 GM117058 (to Y.G.), and NIH R01 HL109810 (to Y.G.). We also thank NIH Shared Instrument Program Grant S10 RR029531 and high-end instrument grant S10 OD018475 for funding the MS instruments.
Author information
Authors and Affiliations
Contributions
J.J. oversaw all aspects of the experiments and manuscript preparation. C.-W.H. performed mass spectrometric, crosslinking, and cell culture experiments. M.W. and H.L. synthesized the compounds. D.F. performed mutagenesis, in-gel fluorescence scanning, and enzyme kinetic experiments. B.L. obtained protein crystals and determined structures. L. Lu assisted with protein purification. X.Z., L. Li, Z.L., L.W., and Y.G. provided access to the MS instruments. C.-W.H. and J.J. wrote the manuscript with help from M.W., D.F., B.L., and H.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–5, Supplementary Figures 1–14 and Supplementary Notes 1–3 (PDF 8347 kb)
Rights and permissions
About this article
Cite this article
Hu, CW., Worth, M., Fan, D. et al. Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase. Nat Chem Biol 13, 1267–1273 (2017). https://doi.org/10.1038/nchembio.2494
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2494
This article is cited by
-
Motif-dependent binding on the intervening domain regulates O-GlcNAc transferase
Nature Chemical Biology (2023)
-
Genetically encoded chemical crosslinking of carbohydrate
Nature Chemistry (2023)
-
Cryo-EM structure provides insights into the dimer arrangement of the O-linked β-N-acetylglucosamine transferase OGT
Nature Communications (2021)
-
Identification of targets of AMPylating Fic enzymes by co-substrate-mediated covalent capture
Nature Chemistry (2020)