Abstract
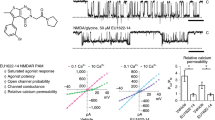
N-Methyl-D-aspartate (NMDA) receptors are the main calcium-permeable excitatory receptors in the mammalian central nervous system. The NMDA receptor gating is complex, exhibiting multiple closed, open, and desensitized states; however, central questions regarding the conformations and energetics of the transmembrane domains as they relate to the gating states are still unanswered. Here, using single-molecule Förster resonance energy transfer (smFRET), we map the energy landscape of the first transmembrane segment of the Rattus norvegicus NMDA receptor under resting and various liganded conditions. These results show kinetically and structurally distinct changes associated with apo, agonist-bound, and inhibited receptors linked by a linear mechanism of gating at this site. Furthermore, the smFRET data suggest that allosteric inhibition by zinc occurs by an uncoupling of the agonist-induced changes at the extracellular domains from the gating motions leading to an apo-like state, while dizocilpine, a pore blocker, stabilizes multiple closely packed transmembrane states.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zhang, W., Howe, J.R. & Popescu, G.K. Distinct gating modes determine the biphasic relaxation of NMDA receptor currents. Nat. Neurosci. 11, 1373–1375 (2008).
Lape, R., Colquhoun, D. & Sivilotti, L.G. On the nature of partial agonism in the nicotinic receptor superfamily. Nature 454, 722–727 (2008).
Zhang, W., Devi, S.P., Tomita, S. & Howe, J.R. Auxiliary proteins promote modal gating of AMPA- and kainate-type glutamate receptors. Eur. J. Neurosci. 39, 1138–1147 (2014).
Tajima, N. et al. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534, 63–68 (2016).
Popescu, G., Robert, A., Howe, J.R. & Auerbach, A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature 430, 790–793 (2004).
Banke, T.G. & Traynelis, S.F. Activation of NR1/NR2B NMDA receptors. Nat. Neurosci. 6, 144–152 (2003).
Kussius, C.L. & Popescu, G.K. Kinetic basis of partial agonism at NMDA receptors. Nat. Neurosci. 12, 1114–1120 (2009).
Zhu, S. et al. Mechanism of NMDA receptor inhibition and activation. Cell 165, 704–714 (2016).
Karakas, E. & Furukawa, H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–997 (2014).
Kim, J.Y., Kim, C. & Lee, N.K. Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering. Nat. Commun. 6, 6992 (2015).
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Sasmal, D.K. & Lu, H.P. Single-molecule patch-clamp FRET microscopy studies of NMDA receptor ion channel dynamics in living cells: revealing the multiple conformational states associated with a channel at its electrical off state. J. Am. Chem. Soc. 136, 12998–13005 (2014).
Monyer, H. et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256, 1217–1221 (1992).
Traynelis, S.F. et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496 (2010).
Kazi, R., Dai, J., Sweeney, C., Zhou, H.-X. & Wollmuth, L.P. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat. Neurosci. 17, 914–922 (2014).
Sobolevsky, A.I., Beck, C. & Wollmuth, L.P. Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron 33, 75–85 (2002).
Borschel, W.F., Cummings, K.A., Tindell, L.K. & Popescu, G.K. Kinetic contributions to gating by interactions unique to N-methyl-D-aspartate (NMDA) receptors. J. Biol. Chem. 290, 26846–26855 (2015).
Amico-Ruvio, S.A., Murthy, S.E., Smith, T.P. & Popescu, G.K. Zinc effects on NMDA receptor gating kinetics. Biophys. J. 100, 1910–1918 (2011).
Sirrieh, R.E., MacLean, D.M. & Jayaraman, V. A conserved structural mechanism of NMDA receptor inhibition: A comparison of ifenprodil and zinc. J. Gen. Physiol. 146, 173–181 (2015).
Sirrieh, R.E., MacLean, D.M. & Jayaraman, V. Amino-terminal domain tetramer organization and structural effects of zinc binding in the N-methyl-D-aspartate (NMDA) receptor. J. Biol. Chem. 288, 22555–22564 (2013).
Sirrieh, R.E., MacLean, D.M. & Jayaraman, V. Subtype-dependent N-methyl-D-aspartate receptor amino-terminal domain conformations and modulation by spermine. J. Biol. Chem. 290, 12812–12820 (2015).
Rambhadran, A., Gonzalez, J. & Jayaraman, V. Conformational changes at the agonist binding domain of the N-methyl-D-aspartic acid receptor. J. Biol. Chem. 286, 16953–16957 (2011).
Cooper, D.R. et al. Conformational transitions in the glycine-bound GluN1 NMDA receptor LBD via single-molecule FRET. Biophys. J. 109, 66–75 (2015).
Dolino, D.M. et al. Structural dynamics of the glycine-binding domain of the N-methyl-D-aspartate receptor. J. Biol. Chem. 290, 797–804 (2015).
Ramaswamy, S. et al. Role of conformational dynamics in α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor partial agonism. J. Biol. Chem. 287, 43557–43564 (2012).
Landes, C.F., Rambhadran, A., Taylor, J.N., Salatan, F. & Jayaraman, V. Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nat. Chem. Biol. 7, 168–173 (2011).
Taylor, J.N., Makarov, D.E. & Landes, C.F. Denoising single-molecule FRET trajectories with wavelets and Bayesian inference. Biophys. J. 98, 164–173 (2010).
Taylor, J.N. & Landes, C.F. Improved resolution of complex single-molecule FRET systems via wavelet shrinkage. J. Phys. Chem. B 115, 1105–1114 (2011).
Shuang, B. et al. Fast step transition and state identification (STaSI) for discrete single-molecule data analysis. J. Phys. Chem. Lett. 5, 3157–3161 (2014).
Chizhik, A.M. et al. Super-resolution optical fluctuation bio-imaging with dual-color carbon nanodots. Nano Lett. 16, 237–242 (2016).
Hu, Z. et al. Excitonic energy migration in conjugated polymers: the critical role of interchain morphology. J. Am. Chem. Soc. 136, 16023–16031 (2014).
Wedeking, T. et al. Single cell GFP-trap reveals stoichiometry and dynamics of cytosolic protein complexes. Nano Lett. 15, 3610–3615 (2015).
Yang, J., Park, H. & Kaufman, L.J. Highly anisotropic conjugated polymer aggregates: preparation and quantification of physical and optical anisotropy. J. Phys. Chem. C 121, 13854–13862 (2017).
Zhang, W., Caldarola, M., Pradhan, B. & Orrit, M. Gold nanorod enhanced fluorescence enables single-molecule electrochemistry of methylene blue. Angew. Chem. Int. Ed. Engl. 56, 3566–3569 (2017).
Alam, A. & Jiang, Y. High-resolution structure of the open NaK channel. Nat. Struct. Mol. Biol. 16, 30–34 (2009).
Shi, N., Ye, S., Alam, A., Chen, L. & Jiang, Y. Atomic structure of a Na+- and K+-conducting channel. Nature 440, 570–574 (2006).
Dolino, D.M., Rezaei Adariani, S., Shaikh, S.A., Jayaraman, V. & Sanabria, H. Conformational selection and submillisecond dynamics of the ligand-binding domain of the N-methyl-D-aspartate receptor. J. Biol. Chem. 291, 16175–16185 (2016).
Yao, Y., Belcher, J., Berger, A.J., Mayer, M.L. & Lau, A. Y. Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure 21, 1788–1799 (2013).
Peters, S., Koh, J. & Choi, D.W. Zinc selectively blocks the action of N-methyl-D-aspartate on cortical neurons. Science 236, 589–593 (1987).
Low, C.M., Zheng, F., Lyuboslavsky, P. & Traynelis, S.F. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-D-aspartate NR1/NR2A receptors. Proc. Natl. Acad. Sci. USA 97, 11062–11067 (2000).
Wong, E.H. et al. The anticonvulsant MK-801 is a potent N-methyl-D-aspartate antagonist. Proc. Natl. Acad. Sci. USA 83, 7104–7108 (1986).
Foster, A.C. & Wong, E.H. The novel anticonvulsant MK-801 binds to the activated state of the N-methyl-D-aspartate receptor in rat brain. Br. J. Pharmacol. 91, 403–409 (1987).
Lü, W., Du, J., Goehring, A. & Gouaux, E. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 355, eaal3729 (2017).
Huettner, J.E. & Bean, B.P. Block of N-methyl-D-aspartate-activated current by the anticonvulsant MK-801: selective binding to open channels. Proc. Natl. Acad. Sci. USA 85, 1307–1311 (1988).
Sobolevsky, A.I. Two-component blocking kinetics of open NMDA channels by organic cations. Biochim. Biophys. Acta 1416, 69–91 (1999).
Sobolevsky, A. & Koshelev, S. Two blocking sites of amino-adamantane derivatives in open N-methyl-D-aspartate channels. Biophys. J. 74, 1305–1319 (1998).
Blanpied, T.A., Boeckman, F.A., Aizenman, E. & Johnson, J.W. Trapping channel block of NMDA-activated responses by amantadine and memantine. J. Neurophysiol. 77, 309–323 (1997).
Mortensen, M. & Smart, T.G. Single-channel recording of ligand-gated ion channels. Nat. Protoc. 2, 2826–2841 (2007).
Wang, J. & Wolynes, P. Instantons and the fluctuating path description of reactions in complex environments. J. Phys. Chem. 100, 1129–1136 (1996).
Pirchi, M. et al. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nat. Commun. 2, 493 (2011).
MacLean, D.M., Ramaswamy, S.S., Du, M., Howe, J.R. & Jayaraman, V. Stargazin promotes closure of the AMPA receptor ligand-binding domain. J. Gen. Physiol. 144, 503–512 (2014).
Shaikh, S.A. et al. Stargazin modulation of AMPA receptors. Cell Rep. 17, 328–335 (2016).
Jain, A. et al. Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488 (2011).
Nick Taylor, J., Darugar, Q., Kourentzi, K., Willson, R.C. & Landes, C.F. Dynamics of an anti-VEGF DNA aptamer: a single-molecule study. Biochem. Biophys. Res. Commun. 373, 213–218 (2008).
Acknowledgements
Methods and additional data can be found in the supplementary materials. This project was supported by NIH grant R35GM122528 to V.J., K99NS094761to D.M.M., American Heart Association Fellowship 16POST30030007 to S.A.S., Schissler Foundation Fellowship to D.M.D., and Welch Foundation Grant C-1787 to C.F.L.
Author information
Authors and Affiliations
Contributions
D.M.D. performed the mutations and prepared the labeled proteins, analyzed the smFRET data, and contributed to designing the research, interpreting the results, and writing the manuscript. S.C. performed the smFRET measurements, analyzed the smFRET data, and contributed to designing the research, interpreting the results, and writing the manuscript. D.M.M. and S.A.S. performed the electrophysiology. C.F. and L.D.C.B. analyzed the smFRET data. C.F.L., and V.J. analyzed the smFRET data and contributed to designing the research, interpreting the results, and writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–7 (PDF 1105 kb)
Rights and permissions
About this article
Cite this article
Dolino, D., Chatterjee, S., MacLean, D. et al. The structure–energy landscape of NMDA receptor gating. Nat Chem Biol 13, 1232–1238 (2017). https://doi.org/10.1038/nchembio.2487
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2487
This article is cited by
-
Conformational rearrangement of the NMDA receptor amino-terminal domain during activation and allosteric modulation
Nature Communications (2021)
-
Zinc as a countermeasure for cadmium toxicity
Acta Pharmacologica Sinica (2021)
-
cAMP binding to closed pacemaker ion channels is non-cooperative
Nature (2021)
-
The structural arrangement at intersubunit interfaces in homomeric kainate receptors
Scientific Reports (2019)