Abstract
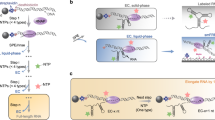
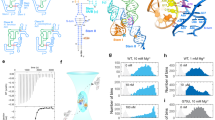
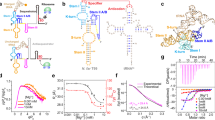
S-adenosyl-L-methionine (SAM) ligand binding induces major structural changes in SAM-I riboswitches, through which gene expression is regulated via transcription termination. Little is known about the conformations and motions governing the function of the full-length Bacillus subtilis yitJ SAM-I riboswitch. Therefore, we have explored its conformational energy landscape as a function of Mg2+ and SAM ligand concentrations using single-molecule Förster resonance energy transfer (smFRET) microscopy and hidden Markov modeling analysis. We resolved four conformational states both in the presence and the absence of SAM and determined their Mg2+-dependent fractional populations and conformational dynamics, including state lifetimes, interconversion rate coefficients and equilibration timescales. Riboswitches with terminator and antiterminator folds coexist, and SAM binding only gradually shifts the populations toward terminator states. We observed a pronounced acceleration of conformational transitions upon SAM binding, which may be crucial for off-switching during the brief decision window before expression of the downstream gene.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Montange, R.K. & Batey, R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441, 1172–1175 (2006).
Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E. & Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10, 701–707 (2003).
Winkler, W.C. & Breaker, R.R. Genetic control by metabolite-binding riboswitches. ChemBioChem 4, 1024–1032 (2003).
Montange, R.K. & Batey, R.T. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 37, 117–133 (2008).
Breaker, R.R. Prospects for riboswitch discovery and analysis. Mol. Cell 43, 867–879 (2011).
Tucker, B.J. & Breaker, R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 15, 342–348 (2005).
Wang, J.X. & Breaker, R.R. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem. Cell Biol. 86, 157–168 (2008).
Trausch, J.J. et al. Structural basis for diversity in the SAM clan of riboswitches. Proc. Natl. Acad. Sci. USA 111, 6624–6629 (2014).
McDaniel, B.A., Grundy, F.J., Artsimovitch, I. & Henkin, T.M. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100, 3083–3088 (2003).
Mustoe, A.M., Brooks, C.L. & Al-Hashimi, H.M. Hierarchy of RNA functional dynamics. Annu. Rev. Biochem. 83, 441–466 (2014).
Fürtig, B., Nozinovic, S., Reining, A. & Schwalbe, H. Multiple conformational states of riboswitches fine-tune gene regulation. Curr. Opin. Struct. Biol. 30, 112–124 (2015).
Lemay, J.F., Penedo, J.C., Tremblay, R., Lilley, D.M. & Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 13, 857–868 (2006).
Greenleaf, W.J., Frieda, K.L., Foster, D.A., Woodside, M.T. & Block, S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319, 630–633 (2008).
Wickiser, J.K., Winkler, W.C., Breaker, R.R. & Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Neupane, K., Yu, H., Foster, D.A., Wang, F. & Woodside, M.T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 39, 7677–7687 (2011).
Rieder, U., Kreutz, C. & Micura, R. Folding of a transcriptionally acting preQ1 riboswitch. Proc. Natl. Acad. Sci. USA 107, 10804–10809 (2010).
Stoddard, C.D. et al. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18, 787–797 (2010).
Montange, R.K. et al. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J. Mol. Biol. 396, 761–772 (2010).
Heppell, B. & Lafontaine, D.A. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry 47, 1490–1499 (2008).
Heppell, B. et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat. Chem. Biol. 7, 384–392 (2011).
Klein, D.J., Schmeing, T.M., Moore, P.B. & Steitz, T.A. The kink-turn: a new RNA secondary structure motif. EMBO J. 20, 4214–4221 (2001).
Winkler, W.C., Grundy, F.J., Murphy, B.A. & Henkin, T.M. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7, 1165–1172 (2001).
McDaniel, B.A., Grundy, F.J. & Henkin, T.M. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol. Microbiol. 57, 1008–1021 (2005).
Lu, C. et al. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J. Mol. Biol. 404, 803–818 (2010).
Aboul-ela, F., Huang, W., Abd Elrahman, M., Boyapati, V. & Li, P. Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design. Wiley Interdiscip Rev RNA 6, 631–650 (2015).
Hennelly, S.P., Novikova, I.V. & Sanbonmatsu, K.Y. The expression platform and the aptamer: cooperativity between Mg2+ and ligand in the SAM-I riboswitch. Nucleic Acids Res. 41, 1922–1935 (2013).
Boyapati, V.K., Huang, W., Spedale, J. & Aboul-Ela, F. Basis for ligand discrimination between ON and OFF state riboswitch conformations: the case of the SAM-I riboswitch. RNA 18, 1230–1243 (2012).
Frieda, K.L. & Block, S.M. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338, 397–400 (2012).
Kurschat, W.C., Müller, J., Wombacher, R. & Helm, M. Optimizing splinted ligation of highly structured small RNAs. RNA 11, 1909–1914 (2005).
Chung, H.S. & Gopich, I.V. Fast single-molecule FRET spectroscopy: theory and experiment. Phys. Chem. Chem. Phys. 16, 18644–18657 (2014).
Dammertz, K., Hengesbach, M., Helm, M., Nienhaus, G.U. & Kobitski, A.Y. Single-molecule FRET studies of counterion effects on the free energy landscape of human mitochondrial lysine tRNA. Biochemistry 50, 3107–3115 (2011).
Kobitski, A.Y., Nierth, A., Helm, M., Jäschke, A. & Nienhaus, G.U. Mg2+-dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Res. 35, 2047–2059 (2007).
Rieger, R., Kobitski, A., Sielaff, H. & Nienhaus, G.U. Evidence of a folding intermediate in RNase H from single-molecule FRET experiments. ChemPhysChem 12, 627–633 (2011).
Hennelly, S.P. & Sanbonmatsu, K.Y. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 39, 2416–2431 (2011).
Pirchi, M. et al. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nat. Commun. 2, 493 (2011).
McKinney, S.A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006).
Lee, T.H. Extracting kinetics information from single-molecule fluorescence resonance energy transfer data using hidden Markov models. J. Phys. Chem. B 113, 11535–11542 (2009).
Keller, B.G., Kobitski, A., Jäschke, A., Nienhaus, G.U. & Noé, F. Complex RNA folding kinetics revealed by single-molecule FRET and hidden Markov models. J. Am. Chem. Soc. 136, 4534–4543 (2014).
Prinz, J.H. et al. Markov models of molecular kinetics: generation and validation. J. Chem. Phys. 134, 174105 (2011).
Huang, W., Kim, J., Jha, S. & Aboul-Ela, F. Conformational heterogeneity of the SAM-I riboswitch transcriptional ON state: a chaperone-like role for S-adenosyl methionine. J. Mol. Biol. 418, 331–349 (2012).
Huang, W., Kim, J., Jha, S. & Aboul-ela, F. The impact of a ligand binding on strand migration in the SAM-I riboswitch. PLoS Comput. Biol. 9, e1003069 (2013).
Reining, A. et al. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature 499, 355–359 (2013).
Frauenfelder, H., Sligar, S.G. & Wolynes, P.G. The energy landscapes and motions of proteins. Science 254, 1598–1603 (1991).
Nienhaus, G.U., Müller, J.D., McMahon, B.H. & Frauenfelder, H. Exploring the conformational energy landscape of proteins. Physica D 107, 297–311 (1997).
Baird, N.J., Kulshina, N. & Ferré-D'Amaré, A.R. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 7, 328–332 (2010).
Suddala, K.C., Wang, J., Hou, Q. & Walter, N.G. Mg2+ shifts ligand-mediated folding of a riboswitch from induced-fit to conformational selection. J. Am. Chem. Soc. 137, 14075–14083 (2015).
Vogel, U. & Jensen, K.F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 176, 2807–2813 (1994).
Huang, W., Kim, J., Jha, S. & Aboul-ela, F. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: a small molecule with a strong arm. Nucleic Acids Res. 37, 6528–6539 (2009).
Whitford, P.C. et al. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys. J. 96, L7–L9 (2009).
Heyes, C.D., Kobitski, A.Y., Amirgoulova, E.V. & Nienhaus, G.U. Biocompatible surfaces for specific tethering of individual protein molecules. J. Phys. Chem. B 108, 13387–13394 (2004).
Aitken, C.E., Marshall, R.A. & Puglisi, J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008).
Dave, R., Terry, D.S., Munro, J.B. & Blanchard, S.C. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009).
Kuzmenkina, E.V., Heyes, C.D. & Nienhaus, G.U. Single-molecule Forster resonance energy transfer study of protein dynamics under denaturing conditions. Proc. Natl. Acad. Sci. USA 102, 15471–15476 (2005).
Seyfried, V., Birk, H., Storz, R. & Ulrich, H. Advances in multispectral confocal imaging. Proc. SPIE 5139, 147–157 (2003).
Kapanidis, A.N. et al. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA 101, 8936–8941 (2004).
Osborne, M.A., Balasubramanian, S., Furey, W.S. & Klenerman, D. Optically biased diffusion of single molecules studied by confocal fluorescence microscopy. J. Phys. Chem. B 102, 3160–3167 (1998).
Saccà, B. et al. Reversible reconfiguration of DNA origami nanochambers monitored by single-molecule FRET. Angew. Chem. Int. Ed. Engl. 54, 3592–3597 (2015).
Kobitski, A.Y., Hengesbach, M., Helm, M. & Nienhaus, G.U. Sculpting an RNA conformational energy landscape by a methyl group modification--a single-molecule FRET study. Angew. Chem. Int. Ed. Engl. 47, 4326–4330 (2008).
Noé, F., Horenko, I., Schütte, C. & Smith, J.C. Hierarchical analysis of conformational dynamics in biomolecules: transition networks of metastable states. J. Chem. Phys. 126, 155102 (2007).
Acknowledgements
We thank A. Schug (KIT) for fruitful discussions. This work was supported by grants from the Karlsruhe Heidelberg Research Partnership (HEiKA) and the Volkswagen Foundation (grant 82549) to A.J. and G.U.N. G.U.N. was also funded by the Helmholtz program Science and Technology of Nanosystems (STN), Karlsruhe School of Optics and Photonics (KSOP) and the Deutsche Forschungsgemeinschaft (DFG), grant GRK 2039. B.G.K. was funded by the DFG through grant CRC 1114 (project B05).
Author information
Authors and Affiliations
Contributions
G.U.N. and A.J. designed research; A.S. synthesized RNA constructs; C.M. and A.Y.K. built the experimental setup and took data; C.M., A.Y.K. and B.G.K. analyzed data and all authors contributed to manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1 and 2, and Supplementary Figures 1–8. (PDF 2215 kb)
Rights and permissions
About this article
Cite this article
Manz, C., Kobitski, A., Samanta, A. et al. Single-molecule FRET reveals the energy landscape of the full-length SAM-I riboswitch. Nat Chem Biol 13, 1172–1178 (2017). https://doi.org/10.1038/nchembio.2476
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2476
This article is cited by
-
Observation of structural switch in nascent SAM-VI riboswitch during transcription at single-nucleotide and single-molecule resolution
Nature Communications (2023)
-
Structure-based insights into recognition and regulation of SAM-sensing riboswitches
Science China Life Sciences (2023)
-
Super-resolution RNA imaging using a rhodamine-binding aptamer with fast exchange kinetics
Nature Biotechnology (2021)
-
Exploring the energy landscape of a SAM-I riboswitch
Journal of Biological Physics (2021)