Abstract
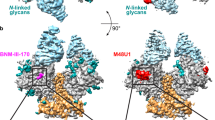
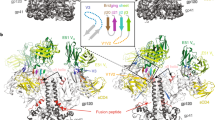
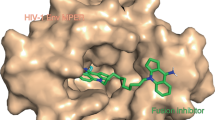
The HIV-1 envelope (Env) spike is a conformational machine that transitions between prefusion (closed, CD4- and CCR5-bound) and postfusion states to facilitate HIV-1 entry into cells. Although the prefusion closed conformation is a potential target for inhibition, development of small-molecule leads has been stymied by difficulties in obtaining structural information. Here, we report crystal structures at 3.8-Å resolution of an HIV-1-Env trimer with BMS-378806 and a derivative BMS-626529 for which a prodrug version is currently in Phase III clinical trials. Both lead candidates recognized an induced binding pocket that was mostly excluded from solvent and comprised of Env elements from a conserved helix and the β20–21 hairpin. In both structures, the β20–21 region assumed a conformation distinct from prefusion-closed and CD4-bound states. Together with biophysical and antigenicity characterizations, the structures illuminate the allosteric and competitive mechanisms by which these small-molecule leads inhibit CD4-induced structural changes in Env.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lima, V.D. et al. Continued improvement in survival among HIV-infected individuals with newer forms of highly active antiretroviral therapy. AIDS 21, 685–692 (2007).
Marcus, J.L. et al. Narrowing the gap in life expectancy between HIV-infected and HIV-uninfected individuals with access to care. J. Acquir. Immune Defic. Syndr. 73, 39–46 (2016).
Vittinghoff, E. et al. Combination antiretroviral therapy and recent declines in AIDS incidence and mortality. J. Infect. Dis. 179, 717–720 (1999).
Kwon, Y.D. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015).
Wyatt, R. & Sodroski, J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 (1998).
Munro, J.B. et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 (2014).
Wild, C., Greenwell, T. & Matthews, T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retroviruses 9, 1051–1053 (1993).
Oldfield, V., Keating, G.M. & Plosker, G. Enfuvirtide: a review of its use in the management of HIV infection. Drugs 65, 1139–1160 (2005).
Dorr, P. et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49, 4721–4732 (2005).
Wu, X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010).
Huang, J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012).
Muster, T. et al. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68, 4031–4034 (1994).
Huang, J. et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121 (2016).
Kwong, P.D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Zhao, Q. et al. Identification of N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology 339, 213–225 (2005).
LaLonde, J.M. et al. Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors. J. Med. Chem. 55, 4382–4396 (2012).
Madani, N. et al. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16, 1689–1701 (2008).
Huang, C.C. et al. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure 13, 755–768 (2005).
Wang, T. et al. Discovery of 4-benzoyl-1-[(4-methoxy-1H- pyrrolo[2,3-b]pyridin-3-yl)oxoacetyl]-2- (R)-methylpiperazine (BMS-378806): a novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J. Med. Chem. 46, 4236–4239 (2003).
Guo, Q. et al. Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions. J. Virol. 77, 10528–10536 (2003).
Ho, H.T. et al. Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J. Virol. 80, 4017–4025 (2006).
Lin, P.F. et al. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc. Natl. Acad. Sci. USA 100, 11013–11018 (2003).
Wang, T. et al. Inhibitors of human immunodeficiency virus type 1 (HIV-1) attachment. 5. An evolution from indole to azaindoles leading to the discovery of 1-(4-benzoylpiperazin-1-yl)-2-(4,7-dimethoxy-1H-pyrrolo[2,3-c]pyridin-3-yl)ethane-1,2-dione (BMS-488043), a drug candidate that demonstrates antiviral activity in HIV-1-infected subjects. J. Med. Chem. 52, 7778–7787 (2009).
Kadow, J., Wang, H.G. & Lin, P.F. Small-molecule HIV-1 gp120 inhibitors to prevent HIV-1 entry: an emerging opportunity for drug development. Curr. Opin. Investig. Drugs 7, 721–726 (2006).
Nowicka-Sans, B. et al. In vitro antiviral characteristics of HIV-1 attachment inhibitor BMS-626529, the active component of the prodrug BMS-663068. Antimicrob. Agents Chemother. 56, 3498–3507 (2012).
Brown, J. et al. Compartmental absorption modeling and site of absorption studies to determine feasibility of an extended-release formulation of an HIV-1 attachment inhibitor phosphate ester prodrug. J. Pharm. Sci. 102, 1742–1751 (2013).
Lalezari, J.P. et al. Safety and efficacy of the HIV-1 attachment inhibitor prodrug BMS-663068 in treatment-experienced individuals: 24 week results of AI438011, a phase 2b, randomised controlled trial. Lancet HIV 2, e427–e437 (2015).
Nettles, R.E. et al. Pharmacodynamics, safety, and pharmacokinetics of BMS-663068, an oral HIV-1 attachment inhibitor in HIV-1-infected subjects. J. Infect. Dis. 206, 1002–1011 (2012).
Ray, N. et al. Prediction of virological response and assessment of resistance emergence to the HIV-1 attachment inhibitor BMS-626529 during 8-day monotherapy with its prodrug BMS-663068. J. Acquir. Immune Defic. Syndr. 64, 7–15 (2013).
Zhou, N. et al. Genotypic correlates of susceptibility to HIV-1 attachment inhibitor BMS-626529, the active agent of the prodrug BMS-663068. J. Antimicrob. Chemother. 69, 573–581 (2014).
Pancera, M. et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014).
Sanders, R.W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Si, Z. et al. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. USA 101, 5036–5041 (2004).
Walker, L.M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Huang, J. et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515, 138–142 (2014).
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103, 8060–8065 (2006).
Madani, N. et al. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J. Virol. 78, 3742–3752 (2004).
Zhou, N. et al. In vivo patterns of resistance to the HIV attachment inhibitor BMS-488043. Antimicrob. Agents Chemother. 55, 729–737 (2011).
Schader, S.M. et al. HIV gp120 H375 is unique to HIV-1 subtype CRF01_AE and confers strong resistance to the entry inhibitor BMS-599793, a candidate microbicide drug. Antimicrob. Agents Chemother. 56, 4257–4267 (2012).
Herschhorn, A. et al. A broad HIV-1 inhibitor blocks envelope glycoprotein transitions critical for entry. Nat. Chem. Biol. 10, 845–852 (2014).
Herschhorn, A. et al. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. MBio 7, e01598–16 (2016).
Pancera, M. et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. USA 107, 1166–1171 (2010).
Langley, D.R. et al. Homology models of the HIV-1 attachment inhibitor BMS-626529 bound to gp120 suggest a unique mechanism of action. Proteins 83, 331–350 (2015).
Kong, R., Tan, J.J., Ma, X.H., Chen, W.Z. & Wang, C.X. Prediction of the binding mode between BMS-378806 and HIV-1 gp120 by docking and molecular dynamics simulation. Biochim. Biophys. Acta 1764, 766–772 (2006).
Teixeira, C., Serradji, N., Maurel, F. & Barbault, F. Docking and 3D-QSAR studies of BMS-806 analogs as HIV-1 gp120 entry inhibitors. Eur. J. Med. Chem. 44, 3524–3532 (2009).
Li, L., Chen, H., Zhao, R.N. & Han, J.G. The investigations on HIV-1 gp120 bound with BMS-488043 by using docking and molecular dynamics simulations. J. Mol. Model. 19, 905–917 (2013).
Liu, Q. et al. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 24, 370–378 (2017).
Guttman, M. et al. CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22, 974–984 (2014).
Da, L.T., Quan, J.M. & Wu, Y.D. Understanding the binding mode and function of BMS-488043 against HIV-1 viral entry. Proteins 79, 1810–1819 (2011).
Shrivastava, I.H., Wendel, K. & LaLonde, J.M. Spontaneous rearrangement of the β20/β21 strands in simulations of unliganded HIV-1 glycoprotein, gp120. Biochemistry 51, 7783–7793 (2012).
Li, M. et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 (2005).
Seaman, M.S. et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84, 1439–1452 (2010).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Weiss, M.S. Global indicators of X-ray data quality. J. Appl. Crystallogr. 34, 130–135 (2001).
Karplus, P.A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Davis, I.W., Murray, L.W., Richardson, J.S. & Richardson, D.C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004).
DeLano, W.L. The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA, 2002).
Acknowledgements
We thank J. Stuckey for assistance with figures, members of Structural Biology and Structural Bioinformatics Core Sections, Vaccine Research Center for discussions and comments on the manuscript. We thank J. Baalwa (University of Alabama at Birmingham), D. Ellenberger (National Cancer Institute, NIH), F. Gao (Duke University), B. Hahn (University of Pennsylvania), K. Hong (Chinese Center for Disease Control and Prevention), J. Kim (The International Vaccine Institute), F. McCutchan (Military HIV Research Program), D. Montefiori (Duke University), L. Morris (National Institute of Communicable Diseases), J. Overbaugh (Fred Hutchison Cancer Research Center), E. Sanders-Buell (Military HIV Research Program), G. Shaw (University of Pennsylvania), R. Swanstrom (University of North Carolina at Chapel Hill), M. Thomson (Instituto de Salud Carlos III), S. Tovanabutra (Military HIV Research Program), C. Williamson (University of Cape Town) and L. Zhang (China Medical University Shenyang) for contributing the HIV-1 envelope plasmids used in our neutralization panel. We thank D. Burton (The Scripps Research Institute) and M. Feinberg (International AIDS Vaccine Initiative) for antibody PGT122 and M. Connors (National Institute of Allergy and Infectious Diseases, NIH) for antibody 35O22 used in structural analysis. Funding was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, the Intramural AIDS Targeted Antiretroviral Program, the National Institute of General Medical Sciences, National Institutes of Health (J.S.), the National Science Foundation MCB-1157506 (E.F.) and the National Institutes of Health GM56550 (E.F.). Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
Author information
Authors and Affiliations
Contributions
M.P. determined the structures with assistance from Y.-T.L., G.-Y.C., D.I.S., D.R.L. and P.D.K. T.B. and G.-Y.C. assisted in resistance analysis. S.N. and A.B.M. performed antigenic analyses. S.O'D., R.T.B., M.K.L. and J.R.M. performed neutralization experiments. A.S. and E.F. performed isothermal titration calorimetry experiments. A.D. expressed proteins, and H.G. and D.I.S. purified proteins. R.R. performed sequence entropy analysis. A.F., A.H., N.M. and J.S. contributed mutagenesis analyses and competition analysis with CD4. M.P. and P.D.K. analyzed the data and wrote the paper, with contributions from J.S. and D.R.L.
Corresponding author
Ethics declarations
Competing interests
D.R.L. owns stock in Bristol-Myers Squibb.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–7 and Supplementary Figures 1–6. (PDF 2811 kb)
Rights and permissions
About this article
Cite this article
Pancera, M., Lai, YT., Bylund, T. et al. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat Chem Biol 13, 1115–1122 (2017). https://doi.org/10.1038/nchembio.2460
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2460
This article is cited by
-
Mechanism of action, resistance, interaction, pharmacokinetics, pharmacodynamics, and safety of fostemsavir
BMC Infectious Diseases (2024)
-
A classification algorithm based on dynamic ensemble selection to predict mutational patterns of the envelope protein in HIV-infected patients
Algorithms for Molecular Biology (2023)
-
Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles
Communications Biology (2023)
-
Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir
Nature Communications (2023)
-
Week 240 Efficacy and Safety of Fostemsavir Plus Optimized Background Therapy in Heavily Treatment-Experienced Adults with HIV-1
Infectious Diseases and Therapy (2023)