Abstract
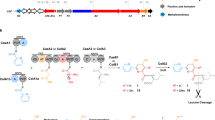
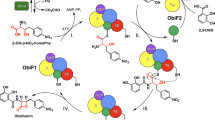
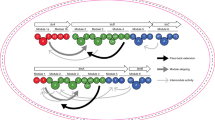
Nonribosomal peptide synthetases (NRPSs) are multidomain modular biosynthetic assembly lines that polymerize amino acids into a myriad of biologically active nonribosomal peptides (NRPs). NRPS thioesterase (TE) domains employ diverse release strategies for off-loading thioester-tethered polymeric peptides from termination modules typically via hydrolysis, aminolysis, or cyclization to provide mature antibiotics as carboxylic acids/esters, amides, and lactams/lactones, respectively. Here we report the enzyme-catalyzed formation of a highly strained β-lactone ring during TE-mediated cyclization of a β-hydroxythioester to release the antibiotic obafluorin (Obi) from an NRPS assembly line. The Obi NRPS (ObiF) contains a type I TE domain with a rare catalytic cysteine residue that plays a direct role in β-lactone ring formation. We present a detailed genetic and biochemical characterization of the entire Obi biosynthetic gene cluster in plant-associated Pseudomonas fluorescens ATCC 39502 that establishes a general strategy for β-lactone biogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lowe, C. & Vederas, J.C. Naturally occuring β-lactones: occurence, synthesis and properties. A review. Org. Prep. Proced. Int. 27, 305–346 (1995).
Bachovchin, D.A. et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. USA 107, 20941–20946 (2010).
Pemble, C.W. IV, Johnson, L.C., Kridel, S.J. & Lowther, W.T. Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nat. Struct. Mol. Biol. 14, 704–709 (2007).
Gulder, T.A. & Moore, B.S. Salinosporamide natural products: Potent 20S proteasome inhibitors as promising cancer chemotherapeutics. Angew. Chem. Int. Edn Engl. 49, 9346–9367 (2010).
De Pascale, G., Nazi, I., Harrison, P.H. & Wright, G.D. β-Lactone natural products and derivatives inactivate homoserine transacetylase, a target for antimicrobial agents. J. Antibiot. (Tokyo) 64, 483–487 (2011).
Lall, M.S., Ramtohul, Y.K., James, M.N.G. & Vederas, J.C. Serine and threonine β-lactones: a new class of hepatitis A virus 3C cysteine proteinase inhibitors. J. Org. Chem. 67, 1536–1547 (2002).
Wyatt, M.A. et al. Biosynthesis of ebelactone A: isotopic tracer, advanced precursor and genetic studies reveal a thioesterase-independent cyclization to give a polyketide β-lactone. J. Antibiot. (Tokyo) 66, 421–430 (2013).
Hamed, R.B. et al. The enzymes of β-lactam biosynthesis. Nat. Prod. Rep. 30, 21–107 (2013).
Roach, P.L. et al. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 387, 827–830 (1997).
Bachmann, B.O., Li, R. & Townsend, C.A. β-Lactam synthetase: a new biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 95, 9082–9086 (1998).
Gaudelli, N.M., Long, D.H. & Townsend, C.A. β-Lactam formation by a non-ribosomal peptide synthetase during antibiotic biosynthesis. Nature 520, 383–387 (2015).
Christenson, J.K. et al. β-Lactone synthetase found in the olefin biosynthesis pathway. Biochemistry 56, 348–351 (2017).
Bai, T. et al. Operon for biosynthesis of lipstatin, the β-lactone inhibitor of human pancreatic lipase. Appl. Environ. Microbiol. 80, 7473–7483 (2014).
Eustáquio, A.S. et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc. Natl. Acad. Sci. USA 106, 12295–12300 (2009).
Zhao, C. et al. Oxazolomycin biosynthesis in Streptomyces albus JA3453 featuring an “acyltransferase-less” type I polyketide synthase that incorporates two distinct extender units. J. Biol. Chem. 285, 20097–20108 (2010).
Horsman, M.E., Hari, T.P. & Boddy, C.N. Polyketide synthase and nonribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate? Nat. Prod. Rep. 33, 183–202 (2016).
Jiang, Y., Morley, K.L., Schrag, J.D. & Kazlauskas, R.J. Different active-site loop orientation in serine hydrolases versus acyltransferases. ChemBioChem 12, 768–776 (2011).
Gaudelli, N.M. & Townsend, C.A. Epimerization and substrate gating by a TE domain in β-lactam antibiotic biosynthesis. Nat. Chem. Biol. 10, 251–258 (2014).
Jensen, K. et al. Polyketide proofreading by an acyltransferase-like enzyme. Chem. Biol. 19, 329–339 (2012).
Kopp, F. & Marahiel, M.A. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat. Prod. Rep. 24, 735–749 (2007).
Dick, L.R. et al. Mechanistic studies on the inactivation of the proteasome by lactacystin: a central role for clasto-lactacystin β-lactone. J. Biol. Chem. 271, 7273–7276 (1996).
Wells, J.S., Trejo, W.H., Principe, P.A. & Sykes, R.B. Obafluorin, a novel β-lactone produced by Pseudomonas fluorescens. Taxonomy, fermentation and biological properties. J. Antibiot. (Tokyo) 37, 802–803 (1984).
Tymiak, A.A., Culver, C.A., Malley, M.F. & Gougoutas, J.Z. Structure of obafluorin: an antibacterial β-lactone from Pseudomonas fluorescens. J. Org. Chem. 50, 5491–5495 (1985).
Dejong, C.A. et al. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol. 12, 1007–1014 (2016).
Hamed, R.B. et al. Crotonase catalysis enables flexible production of functionalized prolines and carbapenams. J. Am. Chem. Soc. 134, 471–479 (2012).
Pu, Y., Lowe, C., Sailer, M. & Vederas, J.C. Synthesis, stability, and antimicrobial activity of (+)-obafluorin and related β-lactone antibiotics. J. Org. Chem. 59, 3642–3655 (1994).
Herbert, R.B. & Knaggs, A.R. Biosynthesis of the antibiotic obafluorin from D-[U-13C]glucose and p-aminophenylalanine in Pseudomonas fluorescens. J. Chem. Soc. Perkin Trans. I 1992, 103–107 (1992).
Herbert, R.B. & Knaggs, A.R. Biosynthesis of the antibiotic obafluorin from p-aminophenylalanine and glycine (glyoxylate). J. Chem. Soc. Perkin Trans. I 1992, 109–113 (1992).
Bentley, R. The shikimate pathway—a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25, 307–384 (1990).
Walsh, C.T., Liu, J., Rusnak, F. & Sakaitani, M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem. Rev. 90, 1105–1129 (1990).
Fernández-Martínez, L.T. et al. New insights into chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712. Antimicrob. Agents Chemother. 58, 7441–7450 (2014).
Blanc, V. et al. Identification and analysis of genes from Streptomyces pristinaespiralis encoding enzymes involved in the biosynthesis of the 4-dimethylamino-L-phenylalanine precursor of pristinamycin I. Mol. Microbiol. 23, 191–202 (1997).
Choi, Y.S., Zhang, H., Brunzelle, J.S., Nair, S.K. & Zhao, H. In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis. Proc. Natl. Acad. Sci. USA 105, 6858–6863 (2008).
Makris, T.M. et al. An unusual peroxo intermediate of the arylamine oxygenase of the chloramphenicol biosynthetic pathway. J. Am. Chem. Soc. 137, 1608–1617 (2015).
Contestabile, R. et al. L-Threonine aldolase, serine hydroxymethyltransferase and fungal alanine racemase. A subgroup of strictly related enzymes specialized for different functions. Eur. J. Biochem. 268, 6508–6525 (2001).
Barnard-Britson, S. et al. Amalgamation of nucleosides and amino acids in antibiotic biosynthesis: discovery of an L-threonine:uridine-5′-aldehyde transaldolase. J. Am. Chem. Soc. 134, 18514–18517 (2012).
Muliandi, A. et al. Biosynthesis of the 4-methyloxazoline-containing nonribosomal peptides, JBIR-34 and -35, in Streptomyces sp. Sp080513GE-23. Chem. Biol. 21, 923–934 (2014).
Zhang, G. et al. Characterization of the amicetin biosynthesis gene cluster from Streptomyces vinaceusdrappus NRRL 2363 implicates two alternative strategies for amide bond formation. Appl. Environ. Microbiol. 78, 2393–2401 (2012).
Reimmann, C., Serino, L., Beyeler, M. & Haas, D. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144, 3135–3148 (1998).
Drake, E.J. et al. Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529, 235–238 (2016).
Röttig, M. et al. NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367 (2011).
Challis, G.L., Ravel, J. & Townsend, C.A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7, 211–224 (2000).
McGrath, N.A. & Raines, R.T. Chemoselectivity in chemical biology: acyl transfer reactions with sulfur and selenium. Acc. Chem. Res. 44, 752–761 (2011).
Li, R., Oliver, R.A. & Townsend, C.A. Identification and characterization of the sulfazecin monobactam biosynthetic gene cluster. Cell Chem. Biol. 24, 24–34 (2017).
Miller, B.R., Drake, E.J., Shi, C., Aldrich, C.C. & Gulick, A.M. Structures of a nonribosomal peptide synthetase module bound to MbtH-like proteins support a highly dynamic domain architecture. J. Biol. Chem. 291, 22559–22571 (2016).
Reimer, J.M., Aloise, M.N., Harrison, P.M. & Schmeing, T.M. Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase. Nature 529, 239–242 (2016).
Ehmann, D.E., Shaw-Reid, C.A., Losey, H.C. & Walsh, C.T. The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates. Proc. Natl. Acad. Sci. USA 97, 2509–2514 (2000).
Smith, S. & Tsai, S.C. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat. Prod. Rep. 24, 1041–1072 (2007).
Kohli, R.M., Takagi, J. & Walsh, C.T. The thioesterase domain from a nonribosomal peptide synthetase as a cyclization catalyst for integrin binding peptides. Proc. Natl. Acad. Sci. USA 99, 1247–1252 (2002).
Makris, T.M., Chakrabarti, M., Münck, E. & Lipscomb, J.D. A family of di-iron monooxygenases catalyzing amino acid β-hydroxylation in antibiotic biosynthesis. Proc. Natl. Acad. Sci. USA 107, 15 391–15396 (2010).
Jiang, W. et al. EcdGHK are three tailoring iron oxygenases for amino acid building blocks of the echinocandin scaffold. J. Am. Chem. Soc. 135, 4457–4466 (2013).
Haslinger, K. et al. The structure of a transient complex of a nonribosomal peptide synthetase and a cytochrome P450 monooxygenase. Angew. Chem. Int. Edn Engl. 53, 8518–8522 (2014).
Chanco, E., Choi, Y.S., Sun, N., Vu, M. & Zhao, H. Characterization of the N-oxygenase AurF from Streptomyces thioletus. Bioorg. Med. Chem. 22, 5569–5577 (2014).
Quadri, L.E. et al. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595 (1998).
Hollenhorst, M.A., Clardy, J. & Walsh, C.T. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl-dipeptide scaffold of the dapdiamide antibiotics. Biochemistry 48, 10467–10472 (2009).
Acknowledgements
This work was supported with start-up funds provided by Washington University in St. Louis and the Research Corporation for Science Advancement through a Cottrell Scholar award to T.A.W. We thank C.T. Walsh (Stanford ChEM-H) for productive scientific discussions. We thank Cofactor Genomics (St. Louis, MO) for bioinformatics consultation and genome sequence analysis. We thank S. Alvarez and B. Evans at the Proteomics & Mass Spectrometry Facility at the Donald Danforth Plant Science Center (St. Louis, MO) for the acquisition of high-resolution MS spectra (NSF Grant No. DBI-0922879). We thank J.-S. Taylor (WUSTL Department of Chemistry) for assistance with the [32P]PPi exchange assay. We thank J. Kao (WUSTL Department of Chemistry) for assistance with NMR experiments.
Author information
Authors and Affiliations
Contributions
T.A.W., J.E.S., and M.R.R. wrote the paper and prepared the supplementary information. T.A.W. oversaw all of the experiments. J.E.S. and M.R.R. cloned and purified ObiG, ObiH, and ObiL. J.E.S. functionally characterized ObiG, ObiH, and ObiL. M.R.R. cloned, purified, and functionally characterized ObiD and ObiF. J.E.S. and M.R.R. purified and characterized all compounds. T.A.W. and N.K.P. isolated Pseudomonas fluorescens gDNA, analyzed sequencing data, and annotated the Obi biosynthetic gene cluster. N.K.P. performed protein homology modeling and helped with preparation of the supplementary information.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–13 and Supplementary Figures 1–23 (PDF 11727 kb)
Supplementary Note
Characterization of Chemical Compounds (PDF 4074 kb)
Rights and permissions
About this article
Cite this article
Schaffer, J., Reck, M., Prasad, N. et al. β-Lactone formation during product release from a nonribosomal peptide synthetase. Nat Chem Biol 13, 737–744 (2017). https://doi.org/10.1038/nchembio.2374
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2374
This article is cited by
-
Characterization of an efficient N-oxygenase from Saccharothrix sp. and its application in the synthesis of azomycin
Biotechnology for Biofuels and Bioproducts (2023)
-
Biosynthesis of a β-lactone natural product globilactone
Nature Synthesis (2023)
-
Discovery of l-threonine transaldolases for enhanced biosynthesis of beta-hydroxylated amino acids
Communications Biology (2023)
-
Discovery and biosynthetic pathway analysis of cyclopentane–β-lactone globilactone A
Nature Synthesis (2023)
-
Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion
Communications Biology (2023)