Abstract
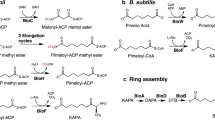
Biotin is an essential vitamin in plants and mammals, functioning as the carbon dioxide carrier within central lipid metabolism. Bacterial pimeloyl-CoA synthetase (BioW) acts as a highly specific substrate-selection gate, ensuring the integrity of the carbon chain in biotin synthesis. BioW catalyzes the condensation of pimelic acid (C7 dicarboxylic acid) with CoASH in an ATP-dependent manner to form pimeloyl-CoA, the first dedicated biotin building block. Multiple structures of Bacillus subtilis BioW together capture all three substrates, as well as the intermediate pimeloyl-adenylate and product pyrophosphate (PPi), indicating that the enzyme uses an internal ruler to select the correct dicarboxylic acid substrate. Both the catalytic mechanism and the surprising stability of the adenylate intermediate were rationalized through site-directed mutagenesis. Building on this understanding, BioW was engineered to synthesize high-value heptanoyl (C7) and octanoyl (C8) monocarboxylic acid-CoA and C8 dicarboxylic-CoA products, highlighting the enzyme's synthetic potential.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Tong, L. Structure and function of biotin-dependent carboxylases. Cell. Mol. Life Sci. 70, 863–891 (2013).
Eisenberg, M.A. The incorporation of 1,7 C14 pimelic acid into biotin vitamers. Biochem. Biophys. Res. Commun. 8, 437–441 (1962).
Eisenberg, M.A. & Krell, K. Synthesis of desthiobiotin from 7,8-diaminopelargonic acid in biotin auxotrophs of Escherichia coli K-12. J. Bacteriol. 98, 1227–1231 (1969).
Eisenberg, M.A. & Star, C. Synthesis of 7-oxo-8-aminopelargonic acid, a biotin vitamer, in cell-free extracts of Escherichia coli biotin auxotrophs. J. Bacteriol. 96, 1291–1297 (1968).
Eisenberg, M.A. & Stoner, G.L. Biosynthesis of 7,8-diaminopelargonic acid, a biotin intermediate, from 7-keto-8-aminopelargonic acid and S-adenosyl-L-methionine. J. Bacteriol. 108, 1135–1140 (1971).
Otsuka, A.J. et al. The Escherichia coli biotin biosynthetic enzyme sequences predicted from the nucleotide sequence of the bio operon. J. Biol. Chem. 263, 19577–19585 (1988).
Lin, S. & Cronan, J.E. Closing in on complete pathways of biotin biosynthesis. Mol. Biosyst. 7, 1811–1821 (2011).
Alexeev, D. et al. The crystal structure of 8-amino-7-oxononanoate synthase: a bacterial PLP-dependent, acyl-CoA-condensing enzyme. J. Mol. Biol. 284, 401–419 (1998).
Alexeev, D., Baxter, R.L. & Sawyer, L. Mechanistic implications and family relationships from the structure of dethiobiotin synthetase. Structure 2, 1061–1072 (1994).
Berkovitch, F., Nicolet, Y., Wan, J.T., Jarrett, J.T. & Drennan, C.L. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 (2004).
Huang, W. et al. Crystal structure of an ATP-dependent carboxylase, dethiobiotin synthetase, at 1.65 Å resolution. Structure 2, 407–414 (1994).
Käck, H., Sandmark, J., Gibson, K., Schneider, G. & Lindqvist, Y. Crystal structure of diaminopelargonic acid synthase: evolutionary relationships between pyridoxal-5′-phosphate-dependent enzymes. J. Mol. Biol. 291, 857–876 (1999).
Webster, S.P. et al. Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies. Biochemistry 39, 516–528 (2000).
Cronan, J.E. & Lin, S. Synthesis of the α,ω-dicarboxylic acid precursor of biotin by the canonical fatty acid biosynthetic pathway. Curr. Opin. Chem. Biol. 15, 407–413 (2011).
Ifuku, O. et al. Origin of carbon atoms of biotin. 13C-NMR studies on biotin biosynthesis in Escherichia coli. Eur. J. Biochem. 220, 585–591 (1994).
Sanyal, I., Lee, S.-L. & Flint, D.H. Biosynthesis of pimeloyl-CoA, a biotin precursor in Escherichia coli, follows a modified fatty acid synthesis pathway: 13C-labelling studies. J. Am. Chem. Soc. 116, 2637–2638 (1994).
Lin, S., Hanson, R.E. & Cronan, J.E. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat. Chem. Biol. 6, 682–688 (2010).
Agarwal, V., Lin, S., Lukk, T., Nair, S.K. & Cronan, J.E. Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis. Proc. Natl. Acad. Sci. USA 109, 17406–17411 (2012).
Bower, S. et al. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J. Bacteriol. 178, 4122–4130 (1996).
Cryle, M.J. & De Voss, J.J. Carbon–carbon bond cleavage by cytochrome p450(BioI)(CYP107H1). Chem. Commun. (Camb.) 2004, 86–87 (2004).
Cryle, M.J. & Schlichting, I. Structural insights from a P450 carrier protein complex reveal how specificity is achieved in the P450(BioI) ACP complex. Proc. Natl. Acad. Sci. USA 105, 15696–15701 (2008).
Gloeckler, R. et al. Cloning and characterization of the Bacillus sphaericus genes controlling the bioconversion of pimelate into dethiobiotin. Gene 87, 63–70 (1990).
Ploux, O., Soularue, P., Marquet, A., Gloeckler, R. & Lemoine, Y. Investigation of the first step of biotin biosynthesis in Bacillus sphaericus. Purification and characterization of the pimeloyl-CoA synthase, and uptake of pimelate. Biochem. J. 287, 685–690 (1992).
Manandhar, M. & Cronan, J.E. Proofreading of noncognate acyl adenylates by an acyl-coenzyme a ligase. Chem. Biol. 20, 1441–1446 (2013).
Gulick, A.M. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 (2009).
Schmelz, S. & Naismith, J.H. Adenylate-forming enzymes. Curr. Opin. Struct. Biol. 19, 666–671 (2009).
Pendini, N.R. et al. Microbial biotin protein ligases aid in understanding holocarboxylase synthetase deficiency. Biochim. Biophys. Acta 1784, 973–982 (2008).
Schmelz, S. et al. AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis. Nat. Chem. Biol. 5, 174–182 (2009).
Finzel, K., Lee, D.J. & Burkart, M.D. Using modern tools to probe the structure–function relationship of fatty acid synthases. ChemBioChem 16, 528–547 (2015).
Upson, R.H., Haugland, R.P., Malekzadeh, M.N. & Haugland, R.P. A spectrophotometric method to measure enzymatic activity in reactions that generate inorganic pyrophosphate. Anal. Biochem. 243, 41–45 (1996).
Webb, M.R. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. USA 89, 4884–4887 (1992).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Yount, R.G., Babcock, D., Ballantyne, W. & Ojala, D. Adenylyl imidodiphosphate, an adenosine triphosphate analog containing a P–N–P linkage. Biochemistry 10, 2484–2489 (1971).
Walzthoeni, T., Leitner, A., Stengel, F. & Aebersold, R. Mass spectrometry supported determination of protein complex structure. Curr. Opin. Struct. Biol. 23, 252–260 (2013).
Choi-Rhee, E., Schulman, H. & Cronan, J.E. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 13, 3043–3050 (2004).
Tron, C.M. et al. Structural and functional studies of the biotin protein ligase from Aquifex aeolicus reveal a critical role for a conserved residue in target specificity. J. Mol. Biol. 387, 129–146 (2009).
Roux, K.J., Kim, D.I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012).
McElroy, W.D., DeLuca, M. & Travis, J. Molecular uniformity in biological catalyses. The enzymes concerned with firefly luciferin, amino acid, and fatty acid utilization are compared. Science 157, 150–160 (1967).
Huang, Y.T., Lu, S.Y., Yi, C.L. & Lee, C.F. Iron-catalyzed synthesis of thioesters from thiols and aldehydes in water. J. Org. Chem. 79, 4561–4568 (2014).
Pal, M. & Bearne, S.L. Synthesis of coenzyme A thioesters using methyl acyl phosphates in an aqueous medium. Org. Biomol. Chem. 12, 9760–9763 (2014).
Beld, J., Finzel, K. & Burkart, M.D. Versatility of acyl-acyl carrier protein synthetases. Chem. Biol. 21, 1293–1299 (2014).
Renata, H., Wang, Z.J. & Arnold, F.H. Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution. Angew. Chem. Int. Edn Engl. 54, 3351–3367 (2015).
Liu, H. & Naismith, J.H. A simple and efficient expression and purification system using two newly constructed vectors. Protein Expr. Purif. 63, 102–111 (2009).
Liu, H. & Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008).
Ploux, O. & Marquet, A. The 8-amino-7-oxopelargonate synthase from Bacillus sphaericus. Purification and preliminary characterization of the cloned enzyme overproduced in Escherichia coli. Biochem. J. 283, 327–331 (1992).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Zhang, Z., Sauter, N.K., van den Bedem, H., Snell, G. & Deacon, A.M. Automated diffraction image analysis and spot searching for high-throughput crystal screening. J. Appl. Crystallogr. 39, 112–119 (2006).
Sauter, N.K., Grosse-Kunstleve, R.W. & Adams, P.D. Robust indexing for automatic data collection. J. Appl. Crystallogr. 37, 399–409 (2004).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Skubák, P. & Pannu, N.S. Automatic protein structure solution from weak X-ray data. Nat. Commun. 4, 2777 (2013).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Terwilliger, T.C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Schüttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC, BB/M003493/1) (D.J.C. and J.H.N.) and Wellcome Trust (WT100209MA (J.H.N.)). The native MS data were acquired on an instrument funded by an Engineering and Physical Sciences Research Council (EPSRC) grant to the University of Edinburgh (EP/K039717/1). We acknowledge the use of the Diamond synchrotron. J.H.N. is a Royal Society Wolfson Merit Award Holder and 1000 talent scholar at Sichuan University. M.W. was funded by a University of Edinburgh PhD scholarship. We thank B. Mykhaylyk for help with bioinformatics analysis and L. Mackay for help with MS analysis.
Author information
Authors and Affiliations
Contributions
A.P. carried out the initial cloning of the bioW gene and characterization of the recombinant BioW. M.W. and P.J.H. carried out the enzyme isolation, characterization and assay. M.W. generated all the BioW mutants and determined the kinetic parameters of wild-type and mutant enzymes. V.K. performed all protein and acyl-CoA MS analyses. M.W. and L.M. prepared the enzyme for crystal trials and optimized crystallization conditions. L.M. carried out the crystallography experiments with J.H.N. and acquired and interpreted the data. L.M., M.W., P.J.H., V.K., J.H.N. and D.J.C. interpreted the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–10 (PDF 4850 kb)
Rights and permissions
About this article
Cite this article
Wang, M., Moynié, L., Harrison, P. et al. Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters. Nat Chem Biol 13, 660–667 (2017). https://doi.org/10.1038/nchembio.2361
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2361
This article is cited by
-
Biochemical and structural characterization of the BioZ enzyme engaged in bacterial biotin synthesis pathway
Nature Communications (2021)
-
Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors
The Journal of Antibiotics (2019)
-
Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme
Nature Chemical Biology (2018)
-
Fatty Acyl-AMP Ligases as Mechanistic Variants of ANL Superfamily and Molecular Determinants Dictating Substrate Specificities
Journal of the Indian Institute of Science (2018)