Abstract
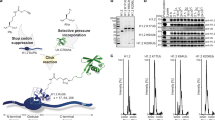
Ubiquitylation of histone H2B, associated with gene activation, leads to chromatin decompaction through an unknown mechanism. We used a hydrogen–deuterium exchange strategy coupled with NMR spectroscopy to map the ubiquitin surface responsible for its structural effects on chromatin. Our studies revealed that a previously uncharacterized acidic patch on ubiquitin comprising residues Glu16 and Glu18 is essential for decompaction. These residues mediate promiscuous electrostatic interactions with the basic histone proteins, potentially positioning the ubiquitin moiety as a dynamic 'wedge' that prevents the intimate association of neighboring nucleosomes. Using two independent crosslinking strategies and an oligomerization assay, we also showed that ubiquitin–ubiquitin contacts occur in the chromatin environment and are important for the solubilization of the chromatin polymers. Our work highlights a novel, chromatin-related aspect of the 'ubiquitin code' and sheds light on how the information-rich ubiquitin modification can orchestrate different biochemical outcomes using distinct surface features.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Woodcock, C.L. & Ghosh, R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2, a000596 (2010).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Sloper-Mould, K.E., Jemc, J.C., Pickart, C.M. & Hicke, L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 276, 30483–30489 (2001).
Dikic, I., Wakatsuki, S. & Walters, K.J. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 (2009).
Braun, S. & Madhani, H.D. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 13, 619–630 (2012).
Wang, H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004).
Zhou, W. et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell 29, 69–80 (2008).
Ng, H.H., Xu, R.M., Zhang, Y. & Struhl, K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277, 34655–34657 (2002).
Sun, Z.W. & Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 (2002).
Minsky, N. et al. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 10, 483–488 (2008).
Kim, J. et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459–471 (2009).
Fuchs, G., Hollander, D., Voichek, Y., Ast, G. & Oren, M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 24, 1572–1583 (2014).
Holt, M.T. et al. Identification of a functional hotspot on ubiquitin required for stimulation of methyltransferase activity on chromatin. Proc. Natl. Acad. Sci. USA 112, 10365–10370 (2015).
Zhou, L. et al. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat. Commun. 7, 10589 (2016).
Fierz, B. et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 7, 113–119 (2011).
Machida, S., Sekine, S., Nishiyama, Y., Horikoshi, N. & Kurumizaka, H. Structural and biochemical analyses of monoubiquitinated human histones H2B and H4. Open Biol. 6, 10.1098/rsob.160090 (2016) .
Paterson, Y., Englander, S.W. & Roder, H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science 249, 755–759 (1990).
Raschke, T.M. & Marqusee, S. Hydrogen exchange studies of protein structure. Curr. Opin. Biotechnol. 9, 80–86 (1998).
Englander, S.W. Protein folding intermediates and pathways studied by hydrogen exchange. Annu. Rev. Biophys. Biomol. Struct. 29, 213–238 (2000).
Zhang, Y.Z., Paterson, Y. & Roder, H. Rapid amide proton exchange rates in peptides and proteins measured by solvent quenching and two-dimensional NMR. Protein Sci. 4, 804–814 (1995).
Hoshino, M. et al. Mapping the core of the beta(2)-microglobulin amyloid fibril by H/D exchange. Nat. Struct. Biol. 9, 332–336 (2002).
Kato, H., Gruschus, J., Ghirlando, R., Tjandra, N. & Bai, Y. Characterization of the N-terminal tail domain of histone H3 in condensed nucleosome arrays by hydrogen exchange and NMR. J. Am. Chem. Soc. 131, 15104–15105 (2009).
Chatterjee, C., McGinty, R.K., Fierz, B. & Muir, T.W. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 6, 267–269 (2010).
Shah, N.H., Dann, G.P., Vila-Perelló, M., Liu, Z. & Muir, T.W. Ultrafast protein splicing is common among cyanobacterial split inteins: implications for protein engineering. J. Am. Chem. Soc. 134, 11338–11341 (2012).
Dyer, P.N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Pan, Y. & Briggs, M.S. Hydrogen exchange in native and alcohol forms of ubiquitin. Biochemistry 31, 11405–11412 (1992).
Going, C.C., Xia, Z. & Williams, E.R. Real-time HD exchange kinetics of proteins from buffered aqueous solution with electrothermal supercharging and top-down tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 27, 1019–1027 (2016).
Schwarz, P.M., Felthauser, A., Fletcher, T.M. & Hansen, J.C. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry 35, 4009–4015 (1996).
Shogren-Knaak, M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Chin, J.W., Martin, A.B., King, D.S., Wang, L. & Schultz, P.G. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. USA 99, 11020–11024 (2002).
Galardy, R.E., Craig, L.C. & Printz, M.P. Benzophenone triplet: a new photochemical probe of biological ligand–receptor interactions. Nat. New Biol. 242, 127–128 (1973).
Song, F. et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380 (2014).
Zheng, C., Lu, X., Hansen, J.C. & Hayes, J.J. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J. Biol. Chem. 280, 33552–33557 (2005).
Nishino, Y. et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 31, 1644–1653 (2012).
Joti, Y. et al. Chromosomes without a 30-nm chromatin fiber. Nucleus 3, 404–410 (2012).
Maeshima, K., Imai, R., Tamura, S. & Nozaki, T. Chromatin as dynamic 10-nm fibers. Chromosoma 123, 225–237 (2014).
Maeshima, K. et al. Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 35, 1115–1132 (2016).
Nakamura, K. et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41, 515–528 (2011).
Moyal, L. et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 41, 529–542 (2011).
Roscoe, B.P., Thayer, K.M., Zeldovich, K.B., Fushman, D. & Bolon, D.N. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J. Mol. Biol. 425, 1363–1377 (2013).
Batta, K., Zhang, Z., Yen, K., Goffman, D.B. & Pugh, B.F. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25, 2254–2265 (2011).
Vijay-Kumar, S., Bugg, C.E. & Cook, W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 194, 531–544 (1987).
Luger, K., Mäder, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Pettersen, E.F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Luger, K., Rechsteiner, T.J., Flaus, A.J., Waye, M.M. & Richmond, T.J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301–311 (1997).
Flaus, A. & Richmond, T.J. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 275, 427–441 (1998).
Farrell, I.S., Toroney, R., Hazen, J.L., Mehl, R.A. & Chin, J.W. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat. Methods 2, 377–384 (2005).
Young, T.S., Ahmad, I., Yin, J.A. & Schultz, P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 395, 361–374 (2010).
Dorigo, B., Schalch, T., Bystricky, K. & Richmond, T.J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Goddard, T.D. & Kneller, D.G. SPARKY 3. (University of California, San Francisco, 2008).
Ueda, T., Chou, H., Kawase, T., Shirakawa, H. & Yoshida, M. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry 43, 9901–9908 (2004).
Acknowledgements
We thank M. Müller, I. Pelczer, B. Fierz, L. Guerra, N. Shah, S. Kilic, M. Holt, U. Nguyen, J. Bos and K. Jani for stimulating discussions and help regarding the preparation of materials. This work was funded by US National Institutes of Health grant R01-GM107047 to T.W.M.
Author information
Authors and Affiliations
Contributions
G.T.D. designed experiments, prepared materials, performed H/D exchange and NMR experiments, compaction assays and crosslinking experiments, analyzed data and wrote the manuscript; K.G. prepared materials, performed compaction assays and analyzed data; T.W.M. designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Figures 1–17. (PDF 8031 kb)
Rights and permissions
About this article
Cite this article
Debelouchina, G., Gerecht, K. & Muir, T. Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat Chem Biol 13, 105–110 (2017). https://doi.org/10.1038/nchembio.2235
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2235
This article is cited by
-
Phosphorylation regulates tau’s phase separation behavior and interactions with chromatin
Communications Biology (2024)
-
RNF8 up-regulates AR/ARV7 action to contribute to advanced prostate cancer progression
Cell Death & Disease (2022)
-
Ultrasound-targeted microbubble destruction-mediated silencing of FBXO11 suppresses development of pancreatic cancer
Human Cell (2022)
-
The solid and liquid states of chromatin
Epigenetics & Chromatin (2021)
-
H1 histones control the epigenetic landscape by local chromatin compaction
Nature (2021)