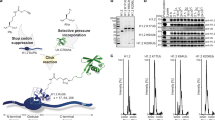
Abstract
Recognition of histone covalent modifications by 'reader' modules constitutes a major mechanism for epigenetic regulation. A recent upsurge of newly discovered histone lysine acylations, such as crotonylation (Kcr), butyrylation (Kbu), and propionylation (Kpr), greatly expands the coding potential of histone lysine modifications. Here we demonstrate that the histone acetylation-binding double PHD finger (DPF) domains of human MOZ (also known as KAT6A) and DPF2 (also known as BAF45d) accommodate a wide range of histone lysine acylations with the strongest preference for Kcr. Crystal structures of the DPF domain of MOZ in complex with H3K14cr, H3K14bu, and H3K14pr peptides reveal that these non-acetyl acylations are anchored in a hydrophobic 'dead-end' pocket with selectivity for crotonylation arising from intimate encapsulation and an amide-sensing hydrogen bonding network. Immunofluorescence and chromatin immunoprecipitation (ChIP)–quantitative PCR (qPCR) showed that MOZ and H3K14cr colocalize in a DPF-dependent manner. Our studies call attention to a new regulatory mechanism centered on histone crotonylation readout by DPF family members.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Strahl, B.D. & Allis, C.D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Taverna, S.D., Li, H., Ruthenburg, A.J., Allis, C.D. & Patel, D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Musselman, C.A., Lalonde, M.E., Côté, J. & Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 (2012).
Patel, D.J. & Wang, Z. Readout of epigenetic modifications. Annu. Rev. Biochem. 82, 81–118 (2013).
Verdin, E. & Ott, M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015).
Huang, H., Sabari, B.R., Garcia, B.A., Allis, C.D. & Zhao, Y. SnapShot: histone modifications. Cell 159, 458–458.e1 (2014).
Huang, H., Lin, S., Garcia, B.A. & Zhao, Y. Quantitative proteomic analysis of histone modifications. Chem. Rev. 115, 2376–2418 (2015).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Montellier, E., Rousseaux, S., Zhao, Y. & Khochbin, S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. BioEssays 34, 187–193 (2012).
Sabari, B.R. et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 (2015).
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999).
Lange, M. et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 22, 2370–2384 (2008).
Zeng, L. et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258–262 (2010).
Li, Y. et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 159, 558–571 (2014).
Li, Y. et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol. Cell 62, 181–193 (2016).
Zhao, D. et al. YEATS2 is a selective histone crotonylation reader. Cell Res. 26, 629–632 (2016).
Andrews, F.H. et al. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 12, 396–398 (2016).
Qiu, Y. et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 26, 1376–1391 (2012).
Ali, M. et al. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J. Mol. Biol. 424, 328–338 (2012).
Dreveny, I. et al. The double PHD finger domain of MOZ/MYST3 induces α-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification. Nucleic Acids Res. 42, 822–835 (2014).
Borrow, J. et al. The translocation t(8;16) (p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 14, 33–41 (1996).
Champagne, N. et al. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J. Biol. Chem. 274, 28528–28536 (1999).
Perez-Campo, F.M., Costa, G., Lie-a-Ling, M., Kouskoff, V. & Lacaud, G. The MYSTerious MOZ, a histone acetyltransferase with a key role in haematopoiesis. Immunology 139, 161–165 (2013).
Yang, X.J. MOZ and MORF acetyltransferases: Molecular interaction, animal development and human disease. Biochim. Biophys. Acta 1853, 1818–1826 (2015).
Wu, J.I. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim. Biophys. Sin. (Shanghai) 44, 54–69 (2012).
Xie, Z. et al. Metabolic Regulation of Gene Expression by Histone Lysine β-Hydroxybutyrylation. Mol. Cell 62, 194–206 (2016).
Arib, G. & Akhtar, A. Multiple facets of nuclear periphery in gene expression control. Curr. Opin. Cell Biol. 23, 346–353 (2011).
Feldman, J.L., Baeza, J. & Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356 (2013).
Bao, X. et al. Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3, 2999 (2014).
Ruthenburg, A.J., Li, H., Patel, D.J. & Allis, C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Li, Y. & Li, H. Many keys to push: diversifying the 'readership' of plant homeodomain fingers. Acta Biochim. Biophys. Sin. (Shanghai) 44, 28–39 (2012).
Li, H. et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28, 677–691 (2007).
Flynn, E.M. et al. A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure 23, 1801–1814 (2015).
Carlson, S. & Glass, K.C. The MOZ histone acetyltransferase in epigenetic signaling and disease. J. Cell. Physiol. 229, 1571–1574 (2014).
Vu, L.P. et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep. 5, 1625–1638 (2013).
Cruickshank, V.A. et al. SWI/SNF Subunits SMARCA4, SMARCD2 and DPF2 Collaborate in MLL-Rearranged Leukaemia Maintenance. PLoS One 10, e0142806 (2015).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Ruthenburg, A.J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Kim, C.H., Kang, M., Kim, H.J., Chatterjee, A. & Schultz, P.G. Site-specific incorporation of ɛ-N-crotonyllysine into histones. Angew. Chem. Int. Edn Engl. 51, 7246–7249 (2012).
Gattner, M.J., Vrabel, M. & Carell, T. Synthesis of ɛ-N-propionyl-, ɛ-N-butyryl-, and ɛ-N-crotonyl-lysine containing histone H3 using the pyrrolysine system. Chem. Commun. (Camb.) 49, 379–381 (2013).
Neumann, H. et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell 36, 153–163 (2009).
Dyer, P.N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Acknowledgements
We thank Z. Chen for critical comments. We thank the staff members at beamline BL17U of the Shanghai Synchrotron Radiation Facility and S. Fan at Tsinghua Center for Structural Biology for their assistance in data collection, and the China National Center for Protein Sciences Beijing for providing facility support. This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0500700 and 2015CB910503), the National Natural Science Foundation of China (91519304), and the Tsinghua University Initiative Scientific Research Program to H.L. We acknowledge support from The Rockefeller University to C.D.A., and the US National Institute of General Medicine to C.D.A. (GM040922), to Y.Z. (GM105933, DK107868, and GM115961), and to T.P. (GM112365).
Author information
Authors and Affiliations
Contributions
H.L. conceived and designed the study; X.X. designed and performed most of the experiments under the guidance of H.L. Y.L. discovered the crotonyllysine reader activity of DPF domain. T.P. performed the designer nucleosome pulldown assay under the guidance of C.D.A. S.Y., P.Y., S.Z. and Y.L. helped with binding and crystallographic experiments. W.Z., W.X., Y.Z. and C.D.A. provided expertise and critical feedback; H.L. and X.X. wrote the manuscript with input from other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–6. (PDF 1783 kb)
Rights and permissions
About this article
Cite this article
Xiong, X., Panchenko, T., Yang, S. et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat Chem Biol 12, 1111–1118 (2016). https://doi.org/10.1038/nchembio.2218
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2218
This article is cited by
-
The mechanisms, regulations, and functions of histone lysine crotonylation
Cell Death Discovery (2024)
-
Epigenetic meets metabolism: novel vulnerabilities to fight cancer
Cell Communication and Signaling (2023)
-
Landscape of mSWI/SNF chromatin remodeling complex perturbations in neurodevelopmental disorders
Nature Genetics (2023)
-
Oncometabolites drive tumorigenesis by enhancing protein acylation: from chromosomal remodelling to nonhistone modification
Journal of Experimental & Clinical Cancer Research (2022)
-
Modulation of cellular processes by histone and non-histone protein acetylation
Nature Reviews Molecular Cell Biology (2022)