Abstract
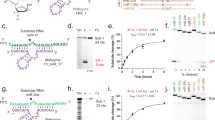
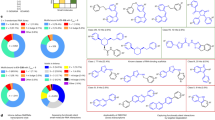
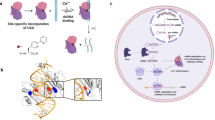
The RNA recognition motif (RRM) is the largest family of eukaryotic RNA-binding proteins. Engineered RRMs with well-defined specificity would provide valuable tools and an exacting test of the current understanding of specificity. We have redesigned the specificity of an RRM using rational methods and demonstrated retargeting of its activity in cells. We engineered the conserved RRM of human Rbfox proteins to specifically bind to the terminal loop of a microRNA precursor (pre-miR-21) with high affinity and inhibit its processing by Drosha and Dicer. We further engineered Giardia Dicer by replacing its PAZ domain with the designed RRM. The reprogrammed enzyme degrades pre-miR-21 specifically in vitro and suppresses mature miR-21 levels in cells, which results in increased expression of the tumor suppressor PDCD4 and significantly decreased viability for cancer cells. The results demonstrate the feasibility of rationally engineering the sequence-specificity of RRMs and of using this ubiquitous platform for diverse biological applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wang, Y., Wang, Z. & Tanaka Hall, T.M. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 280, 3755–3767 (2013).
Mackay, J.P., Font, J. & Segal, D.J. The prospects for designer single-stranded RNA-binding proteins. Nat. Struct. Mol. Biol. 18, 256–261 (2011).
Chen, Y. & Varani, G. Engineering RNA-binding proteins for biology. FEBS J. 280, 3734–3754 (2013).
Cheong, C.G. & Hall, T.M. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. USA 103, 13635–13639 (2006).
Filipovska, A., Razif, M.F.M., Nygård, K.K.A. & Rackham, O. A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 7, 425–427 (2011).
Agrawal, A.A., McLaughlin, K.J., Jenkins, J.L. & Kielkopf, C.L. Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA. Proc. Natl. Acad. Sci. USA 111, 17420–17425 (2014).
Blakeley, B.D. & McNaughton, B.R. Synthetic RNA recognition motifs that selectively recognize HIV-1 trans-activation response element hairpin RNA. ACS Chem. Biol. 9, 1320–1329 (2014).
Kuroyanagi, H. Fox-1 family of RNA-binding proteins. Cell. Mol. Life Sci. 66, 3895–3907 (2009).
Auweter, S.D. et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 25, 163–173 (2006).
Zhang, X. & Zeng, Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 38, 7689–7697 (2010).
Gu, S. et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 151, 900–911 (2012).
Choudhury, N.R. & Michlewski, G. Terminal loop-mediated control of microRNA biogenesis. Biochem. Soc. Trans. 40, 789–793 (2012).
Michlewski, G., Guil, S., Semple, C.A. & Cáceres, J.F. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell 32, 383–393 (2008).
Nam, Y., Chen, C., Gregory, R.I., Chou, J.J. & Sliz, P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080–1091 (2011).
Trabucchi, M. et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459, 1010–1014 (2009).
Castilla-Llorente, V., Nicastro, G. & Ramos, A. Terminal loop-mediated regulation of miRNA biogenesis: selectivity and mechanisms. Biochem. Soc. Trans. 41, 861–865 (2013).
Krichevsky, A.M. & Gabriely, G. miR-21: a small multi-faceted RNA. J. Cell. Mol. Med. 13, 39–53 (2009).
Medina, P.P., Nolde, M. & Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467, 86–90 (2010).
Macrae, I.J. et al. Structural basis for double-stranded RNA processing by Dicer. Science 311, 195–198 (2006).
Asangani, I.A. et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 (2008).
Davis, B.N., Hilyard, A.C., Lagna, G. & Hata, A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56–61 (2008).
Oubridge, C., Ito, N., Evans, P.R., Teo, C.-H. & Nagai, K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372, 432–438 (1994).
Allers, J. & Shamoo, Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 311, 75–86 (2001).
Chen, Y., Kortemme, T., Robertson, T., Baker, D. & Varani, G. A new hydrogen-bonding potential for the design of protein-RNA interactions predicts specific contacts and discriminates decoys. Nucleic Acids Res. 32, 5147–5162 (2004).
Minuchehr, Z. & Goliaei, B. Propensity of amino acids in loop regions connecting β-strands. Protein Pept. Lett. 12, 379–382 (2005).
Weeks, K.M. & Mauger, D.M. Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 44, 1280–1291 (2011).
Chen, Y. et al. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 44, 4381–4395 (2016).
Krol, J., Loedige, I. & Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 (2010).
Nguyen, T.A. et al. Functional anatomy of the human microprocessor. Cell 161, 1374–1387 (2015).
Newman, M.A., Thomson, J.M. & Hammond, S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549 (2008).
MacRae, I.J., Zhou, K. & Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 14, 934–940 (2007).
Peroutka, R.J., Elshourbagy, N., Piech, T. & Butt, T.R. Enhanced protein expression in mammalian cells using engineered SUMO fusions: secreted phospholipase A2. Protein Sci. 17, 1586–1595 (2008).
Si, M.-L. miR-21-mediated tumor growth. Oncogene 26, 2799–2803 (2007).
Schmittgen, T.D., Jiang, J., Liu, Q. & Yang, L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 32, e43 (2004).
Lunde, B.M., Moore, C. & Varani, G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 (2007).
Auweter, S.D., Oberstrass, F.C. & Allain, F.H.-T. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 34, 4943–4959 (2006).
Lünse, C.E. et al. An aptamer targeting the apical-loop domain modulates pri-miRNA processing. Angew. Chem. Int. Ed. Engl. 49, 4674–4677 (2010).
Diaz, J.P. et al. Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by Drosha. RNA 20, 528–539 (2014).
Griffiths-Jones, S., Saini, H.K., van Dongen, S. & Enright, A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158 (2008).
Price, S.R., Oubridge, C., Varani, G. & Nagai, K. Preparation of RNA–protein complexes for X-ray crystallography and NMR. in RNA–Protein Interaction: Practical Approach (ed. Smith, C.) 37–74 (Oxford University Press, 1998).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Wilkinson, K.A., Merino, E.J. & Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 (2006).
Das, R., Laederach, A., Pearlman, S.M., Herschlag, D. & Altman, R.B. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11, 344–354 (2005).
Parisien, M. & Major, F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452, 51–55 (2008).
Tenzer, S. et al. Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J. Proteome Res. 12, 2869–2884 (2013).
Zheng, S., Robertson, T.A. & Varani, G. A knowledge-based potential function predicts the specificity and relative binding energy of RNA-binding proteins. FEBS J. 274, 6378–6391 (2007).
Kortemme, T., Morozov, A.V. & Baker, D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 326, 1239–1259 (2003).
Acknowledgements
The authors are grateful to J. Mandic and other members of G.V.'s laboratory for technical assistance. We thank the Analytical Biopharmacy Core for access to ITC in the School of Pharmacy at the University of Washington. This work was supported by US National Institutes of Health Grant 1R01 GM103834 to G.V., the University of Trento (Progetto Biotecnologie, P.M.) and the Autonomous Province of Trento (Madelena Project, P.M.).
Author information
Authors and Affiliations
Contributions
Y.C. conceived the project and designed the experiments, performed protein design, biochemical and cell-based assays, analyzed the data and wrote the paper; F.Y. performed ITC and NMR analysis of protein–RNA interactions; L.Z. and T.P. performed the cell biological assays; W.Y. cloned and expressed G Dicer proteins; K.G. performed SHAPE analysis; M.W. performed Dicer processing assays. S.Z. performed protein structural modeling; L.Z. and P.M. analyzed the data and wrote the paper; G.V. conceived the project, analyzed the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–7. (PDF 3347 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Yang, F., Zubovic, L. et al. Targeted inhibition of oncogenic miR-21 maturation with designed RNA-binding proteins. Nat Chem Biol 12, 717–723 (2016). https://doi.org/10.1038/nchembio.2128
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2128
This article is cited by
-
Two distinct binding modes provide the RNA-binding protein RbFox with extraordinary sequence specificity
Nature Communications (2023)
-
The kinetic landscape of an RNA-binding protein in cells
Nature (2021)
-
Expanding the binding specificity for RNA recognition by a PUF domain
Nature Communications (2021)
-
Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer
Cellular and Molecular Life Sciences (2019)