Abstract
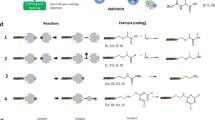
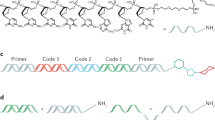
Biochemical combinatorial techniques such as phage display, RNA display and oligonucleotide aptamers have proven to be reliable methods for generation of ligands to protein targets. Adapting these techniques to small synthetic molecules has been a long-sought goal. We report the synthesis and interrogation of an 800-million-member DNA-encoded library in which small molecules are covalently attached to an encoding oligonucleotide. The library was assembled by a combination of chemical and enzymatic synthesis, and interrogated by affinity selection. We describe methods for the selection and deconvolution of the chemical display library, and the discovery of inhibitors for two enzymes: Aurora A kinase and p38 MAP kinase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
17 September 2009
In the version of this article initially published, the IC50 values in the table in Figure 3c were listed as nM instead of μM. The error has been corrected in the HTML and PDF versions of the article.
References
Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Barbas, C.F. III. Recent advances in phage display. Curr. Opin. Biotechnol. 4, 526–530 (1993).
Wilson, D.S., Keefe, A.D. & Szostak, J.W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. USA 98, 3750–3755 (2001).
Frankel, A., Li, S., Starck, S.R. & Roberts, R.W. Unnatural RNA display libraries. Curr. Opin. Struct. Biol. 13, 506–512 (2003).
Ellington, A.D. & Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Joyce, G.F. Amplification, mutation and selection of catalytic RNA. Gene 82, 83–87 (1989).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Brenner, S. & Lerner, R.A. Encoded combinatorial chemistry. Proc. Natl. Acad. Sci. USA 89, 5381–5383 (1992).
Melkko, S., Dumelin, C.E., Scheuermann, J. & Neri, D. Lead discovery by DNA-encoded chemical libraries. Drug Discov. Today 12, 465–471 (2007).
Li, X. & Liu, D.R. DNA-templated organic synthesis: nature's strategy for controlling chemical reactivity applied to synthetic molecules. Angew. Chem. Int. Ed. 43, 4848–4870 (2004).
Halpin, D.R., Lee, J.A., Wrenn, S.J. & Harbury, P.B. DNA display. III. Solid-phase organic synthesis on unprotected DNA. PLoS Biol. 2, 1031–1038 (2004).
Wrenn, S.J., Weisinger, R.W., Halpin, D.R. & Harbury, P.B. Synthetic ligands discovered by in vitro selection. J. Am. Chem. Soc. 129, 13137–13143 (2007).
Melkko, S., Scheurmann, J., Dumelin, C.E. & Neri, D. Encoded self-assembling chemical libraries. Nat. Biotechnol. 22, 568–574 (2004).
Buller, F. et al. Design and synthesis of a novel DNA-encoded chemical library using Diels-Alder cycloadditions. Bioorg. Med. Chem. Lett. 18, 5926–5931 (2008).
Hansen, M.H. et al. A Yoctoliter-scale DNA reactor for small-molecule evolution. J. Am. Chem. Soc. 131, 1322–1327 (2009).
Morgan, B. et al. Synthesis of combinatorial libraries containing encoding oligonucleotide tags. WO patent application 2007/053358 (2007).
Kinoshita, Y. & Nishigaki, K. Enzymic synthesis of code regions for encoded combinational chemistry (ECC). in Nucleic Acids Symposium Series No. 34, 201–202 (Oxford University Press, Oxford, 1995).
Leftheris, K. et al. The discovery of orally active triaminotriazine aniline amides as inhibitors of p38 MAP kinase. J. Med. Chem. 47, 6283–6291 (2004).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Mannoci, L. et al. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc. Natl. Acad. Sci. USA 105, 17670–17675 (2008).
Harrington, E. et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 10, 262–267 (2004).
Mortlock, A.A. et al. Progress in the development of selective inhibitors of Aurora Kinases. Curr. Top. Med. Chem. 5, 807–821 (2005).
Burns, C.J., Wilks, A.F. & Bu, X. Preparation of pyrazine derivatives as kinase inhibitors. WO patent 2005/054230 (2005).
Western, E.C., Daft, J.R., Johnson, E.M., II, Gannett, P.M. & Shaughnessy, K.H. Efficient one-step Suzuki arylation of unprotected halonucleosides, using water-soluble palladium catalysts. J. Org. Chem. 68, 6767–6774 (2003).
Gartner, Z.J., Kanan, M.W. & Liu, D.R. Expanding the reaction scope of DNA-templated synthesis. Angew. Chem. Int. Ed. 41, 1796–1800 (2002).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Methods (PDF 2038 kb)
Supplementary Dataset 1
DEL-A Synthon List (XLS 124 kb)
Supplementary Dataset 2
DEL-B Synthon List (XLS 195 kb)
Supplementary Dataset 3
DEL-A Selection - p38 MAPK (XLS 547 kb)
Supplementary Dataset 4
DEL-A Selection - Aurora A, Method A (XLS 31 kb)
Supplementary Dataset 5
DEL-A Selection - Aurora A, Method B (XLS 20 kb)
Supplementary Dataset 6
DEL-B Selection - p38 MAPK (XLS 24 kb)
Rights and permissions
About this article
Cite this article
Clark, M., Acharya, R., Arico-Muendel, C. et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol 5, 647–654 (2009). https://doi.org/10.1038/nchembio.211
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.211
This article is cited by
-
Protein-templated ligand discovery via the selection of DNA-encoded dynamic libraries
Nature Chemistry (2024)
-
DNA-encoded library-enabled discovery of proximity-inducing small molecules
Nature Chemical Biology (2024)
-
Small-molecule discovery through DNA-encoded libraries
Nature Reviews Drug Discovery (2023)
-
Trio-pharmacophore DNA-encoded chemical library for simultaneous selection of fragments and linkers
Nature Communications (2023)
-
A DNA-encoded chemical library based on chiral 4-amino-proline enables stereospecific isozyme-selective protein recognition
Nature Chemistry (2023)