Abstract
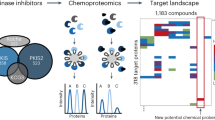
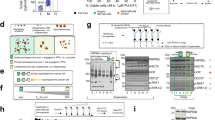
Protein-protein interactions (PPIs) are emerging as a promising new class of drug targets. Here, we present a novel high-throughput approach to screen inhibitors of PPIs in cells. We designed a library of 50,000 human peptide-binding motifs and used a pooled lentiviral system to express them intracellularly and screen for their effects on cell proliferation. We thereby identified inhibitors that drastically reduced the viability of a pancreatic cancer line (RWP1) while leaving a control line virtually unaffected. We identified their target interactions computationally, and validated a subset in experiments. We also discovered their potential mechanisms of action, including apoptosis and cell cycle arrest. Finally, we confirmed that synthetic lipopeptide versions of our inhibitors have similarly specific and dosage-dependent effects on cancer cell growth. Our screen reveals new drug targets and peptide drug leads, and it provides a rich data set covering phenotypes for the inhibition of thousands of interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Booth, B. & Zemmel, R. Prospects for productivity. Nat. Rev. Drug Discov. 3, 451–456 (2004).
Overington, J.P., Al-Lazikani, B. & Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Grant, S.K. Therapeutic protein kinase inhibitors. Cell. Mol. Life Sci. 66, 1163–1177 (2009).
Mullard, A. Protein-protein interaction inhibitors get into the groove. Nat. Rev. Drug Discov. 11, 173–175 (2012).
Fuller, J.C., Burgoyne, N.J. & Jackson, R.M. Predicting druggable binding sites at the protein-protein interface. Drug Discov. Today 14, 155–161 (2009).
Nero, T.L., Morton, C.J., Holien, J.K., Wielens, J. & Parker, M.W. Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer 14, 248–262 (2014).
van Delft, M.F. et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10, 389–399 (2006).
Souers, A.J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
Liu, M. et al. D-peptide inhibitors of the p53–MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc. Natl. Acad. Sci. USA 107, 14321–14326 (2010).
Shangary, S. & Wang, S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu. Rev. Pharmacol. Toxicol. 49, 223–241 (2009).
Chang, Y.S. et al. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. USA 110, E3445–E3454 (2013).
Khoo, K.H., Verma, C.S. & Lane, D.P. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13, 217–236 (2014).
Neduva, V. et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 3, e405 (2005).
Neduva, V. & Russell, R.B. Linear motifs: evolutionary interaction switches. FEBS Lett. 579, 3342–3345 (2005).
Sims, D. et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol. 12, R104 (2011).
Amado, R.G. & Chen, I.S. Lentiviral vectors–the promise of gene therapy within reach? Science 285, 674–676 (1999).
Ivarsson, Y. et al. Large-scale interaction profiling of PDZ domains through proteomic peptide-phage display using human and viral phage peptidomes. Proc. Natl. Acad. Sci. USA 111, 2542–2547 (2014).
Natarajan, V. et al. Peptides genetically selected for NF-κB activation cooperate with oncogene Ras and model carcinogenic role of inflammation. Proc. Natl. Acad. Sci. USA 111, E474–E483 (2014).
Poyurovsky, M.V. et al. The C terminus of p53 binds the N-terminal domain of MDM2. Nat. Struct. Mol. Biol. 17, 982–989 (2010).
Chène, P. et al. A small synthetic peptide, which inhibits the p53-hdm2 interaction, stimulates the p53 pathway in tumour cell lines. J. Mol. Biol. 299, 245–253 (2000).
Bossi, A. & Lehner, B. Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 5, 260 (2009).
Dinkel, H. et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 42, D259–D266 (2014).
Finn, R.D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Wazir, U., Jiang, W.G., Sharma, A.K. & Mokbel, K. Guanine nucleotide binding protein β 1: a novel transduction protein with a possible role in human breast cancer. Cancer Genomics Proteomics 10, 69–73 (2013).
Barabási, A.L., Gulbahce, N. & Loscalzo, J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 (2011).
Zhong, Q. et al. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 5, 321 (2009).
Sekar, R.B. & Periasamy, A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 160, 629–633 (2003).
Chen, J., Wu, X., Lin, J. & Levine, A.J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol. Cell. Biol. 16, 2445–2452 (1996).
Hermeking, H. et al. 14–3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1, 3–11 (1997).
Rajagopalan, S., Jaulent, A.M., Wells, M., Veprintsev, D.B. & Fersht, A.R. 14–3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res. 36, 5983–5991 (2008).
Masters, S.C. & Fu, H. 14–3-3 proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 276, 45193–45200 (2001).
Wang, B. et al. Isolation of high-affinity peptide antagonists of 14–3-3 proteins by phage display. Biochemistry 38, 12499–12504 (1999).
Ito, K., Caramori, G. & Adcock, I.M. Therapeutic potential of phosphatidylinositol 3-kinase inhibitors in inflammatory respiratory disease. J. Pharmacol. Exp. Ther. 321, 1–8 (2007).
Pumiglia, K.M. et al. A direct interaction between G-protein beta gamma subunits and the Raf-1 protein kinase. J. Biol. Chem. 270, 14251–14254 (1995).
Yoda, A. et al. Novel oncogenic mutations in the beta subunit of heteromeric G-proteins identified by functional cDNA library screening. Mol. Cancer Ther. 12 (suppl. 11), PR07 (2013).
Lamers, F. et al. Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocr. Relat. Cancer 18, 657–668 (2011).
Zhou, M. et al. DNA damage induces a novel p53-survivin signaling pathway regulating cell cycle and apoptosis in acute lymphoblastic leukemia cells. J. Pharmacol. Exp. Ther. 303, 124–131 (2002).
Abraham, J. et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 22, 6137–6147 (2003).
Pamidi, A. et al. Functional interplay of p53 and Mus81 in DNA damage responses and cancer. Cancer Res. 67, 8527–8535 (2007).
Zhang, L. & Bulaj, G. Converting peptides into drug leads by lipidation. Curr. Med. Chem. 19, 1602–1618 (2012).
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996).
Zufferey, R., Nagy, D., Mandel, R.J., Naldini, L. & Trono, D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15, 871–875 (1997).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Marcotte, R. et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2, 172–189 (2012).
Higgins, M.E., Claremont, M., Major, J.E., Sander, C. & Lash, A.E. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 35, D721–D726 (2007).
Berman, H.M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Kanehisa, M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014).
Wallrabe, H., Elangovan, M., Burchard, A., Periasamy, A. & Barroso, M. Confocal FRET microscopy to measure clustering of ligand-receptor complexes in endocytic membranes. Biophys. J. 85, 559–571 (2003).
Collins, J.M., Porter, K.A., Singh, S.K. & Vanier, G.S. High-efficiency solid phase peptide synthesis (HE-SPPS). Org. Lett. 16, 940–943 (2014).
Acknowledgements
We thank the members of the Moffat laboratory for valuable technical assistance with lentiviral screening technology, usage of reagents and equipment. We thank A. Emili, T. Hughes and M. Garton for helpful comments on the manuscript. We thank A. Musacchio (Department of Mechanistic Cell Biology, Max Planck Institute of Molecular Physiology) for providing us with the INCENP cDNA clone. We thank F. Sicheri (Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto) for use of the ITC machine. P.M.K. acknowledges an Operating Grant from the Canadian Institute of Health Research (CIHR MOP-123526) and an Innovation Grant from the Canadian Cancer Society Research Institute (CCSRI# 702884). J.M. is a Tier 2 Canada Research Chair in Functional Genomics of Cancer. The research was supported in part by the Intramural Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (N.I.T.).
Author information
Authors and Affiliations
Contributions
P.M.K. designed the project, provided study guidance and wrote the bulk of the manuscript. S.N. performed most experiments and contributed to writing of the manuscript. J.J. performed all bioinformatics analysis and interpreted results as well as assisting in manuscript preparation. C.C.-V. and M.-H.S. performed affinity measurements and helped with other biochemical experiments. Y.I. provided the oligonucleotide library and provided study guidance. N.T. provided synthetic peptides and guidance on their use. J.M. helped design the project and provided guidance on lentiviral screening.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–11. (PDF 2854 kb)
Supplementary Data Set 1
The list of human peptides for dropout screen and their effect on cell viability. (XLS 12194 kb)
Supplementary Data Set 2
A list of cancer-specific inhibitors. (XLS 217 kb)
Supplementary Data Set 3
Peptide-target interactions (XLS 301 kb)
Rights and permissions
About this article
Cite this article
Nim, S., Jeon, J., Corbi-Verge, C. et al. Pooled screening for antiproliferative inhibitors of protein-protein interactions. Nat Chem Biol 12, 275–281 (2016). https://doi.org/10.1038/nchembio.2026
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2026
This article is cited by
-
Disrupting the α-synuclein-ESCRT interaction with a peptide inhibitor mitigates neurodegeneration in preclinical models of Parkinson’s disease
Nature Communications (2023)
-
Genome‑wide identification of CAMTA gene family members in rice (Oryza sativa L.) and in silico study on their versatility in respect to gene expression and promoter structure
Functional & Integrative Genomics (2022)
-
Development of R7BP inhibitors through cross-linking coupled mass spectrometry and integrated modeling
Communications Biology (2019)
-
Viral journeys on the intracellular highways
Cellular and Molecular Life Sciences (2018)
-
Specific and intrinsic sequence patterns extracted by deep learning from intra-protein binding and non-binding peptide fragments
Scientific Reports (2017)