Abstract
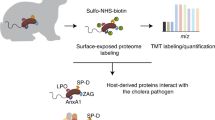
Activity-based protein profiling (ABPP) is a chemoproteomic tool for detecting active enzymes in complex biological systems. We used ABPP to identify secreted bacterial and host serine hydrolases that are active in animals infected with the cholera pathogen Vibrio cholerae. Four V. cholerae proteases were consistently active in infected rabbits, and one, VC0157 (renamed IvaP), was also active in human choleric stool. Inactivation of IvaP influenced the activity of other secreted V. cholerae and rabbit enzymes in vivo, and genetic disruption of all four proteases increased the abundance of intelectin, an intestinal lectin, and its binding to V. cholerae in infected rabbits. Intelectin also bound to other enteric bacterial pathogens, suggesting that it may constitute a previously unrecognized mechanism of bacterial surveillance in the intestine that is inhibited by pathogen-secreted proteases. Our work demonstrates the power of activity-based proteomics to reveal host-pathogen enzymatic dialog in an animal model of infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
23 March 2016
Within the Discussion section, one instance referring to the published crystal structure of trimeric human intelectin-1 was attributed to reference 47 instead of the correct reference 44. The error has been corrected in the HTML and PDF versions of the article.
References
Ivarsson, M.E., Leroux, J.C. & Castagner, B. Targeting bacterial toxins. Angew. Chem. Int. Edn. Engl. 51, 4024–4045 (2012).
Kilian, M., Mestecky, J. & Russell, M.W. Defense mechanisms involving Fc-dependent functions of immunoglobulin A and their subversion by bacterial immunoglobulin A proteases. Microbiol. Rev. 52, 296–303 (1988).
Ruiz-Perez, F. & Nataro, J.P. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell. Mol. Life Sci. 71, 745–770 (2014).
Cravatt, B.F., Wright, A.T. & Kozarich, J.W. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 (2008).
Puri, A.W. & Bogyo, M. Applications of small molecule probes in dissecting mechanisms of bacterial virulence and host responses. Biochemistry 52, 5985–5996 (2013).
Ritchie, J.M. & Waldor, M.K. Vibrio cholerae interactions with the gastrointestinal tract: lessons from animal studies. Curr. Top. Microbiol. Immunol. 337, 37–59 (2009).
Sikora, A.E. Proteins secreted via the type II secretion system: smart strategies of Vibrio cholerae to maintain fitness in different ecological niches. PLoS Pathog. 9, e1003126 (2013).
Simon, G.M. & Cravatt, B.F. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J. Biol. Chem. 285, 11051–11055 (2010).
Kidd, D., Liu, Y. & Cravatt, B.F. Profiling serine hydrolase activities in complex proteomes. Biochemistry 40, 4005–4015 (2001).
Patricelli, M.P., Giang, D.K., Stamp, L.M. & Burbaum, J.J. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics 1, 1067–1071 (2001).
Shaw, D.K., Hyde, J.A. & Skare, J.T. The BB0646 protein demonstrates lipase and haemolytic activity associated with Borrelia burgdorferi, the aetiological agent of Lyme disease. Mol. Microbiol. 83, 319–334 (2012).
Weadge, J.T. & Clarke, A.J. Neisseria gonorrheae O-acetylpeptidoglycan esterase, a serine esterase with a Ser-His-Asp catalytic triad. Biochemistry 46, 4932–4941 (2007).
Stroud, R.M., Kossiakoff, A.A. & Chambers, J.L. Mechanisms of zymogen activation. Annu. Rev. Biophys. Bioeng. 6, 177–193 (1977).
Gloeckl, S. et al. Identification of a serine protease inhibitor which causes inclusion vacuole reduction and is lethal to Chlamydia trachomatis. Mol. Microbiol. 89, 676–689 (2013).
Hauske, P. et al. Selectivity profiling of DegP substrates and inhibitors. Bioorg. Med. Chem. 17, 2920–2924 (2009).
Steuber, H. & Hilgenfeld, R. Recent advances in targeting viral proteases for the discovery of novel antivirals. Curr. Top. Med. Chem. 10, 323–345 (2010).
Tsuji, S. et al. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J. Biol. Chem. 276, 23456–23463 (2001).
Ritchie, J.M., Rui, H., Bronson, R.T. & Waldor, M.K. Back to the future: studying cholera pathogenesis using infant rabbits. MBio 1, e00047–10 (2010).
Bachovchin, D.A. et al. A high-throughput, multiplexed assay for superfamily-wide profiling of enzyme activity. Nat. Chem. Biol. 10, 656–663 (2014).
Weerapana, E., Speers, A.E. & Cravatt, B.F. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2, 1414–1425 (2007).
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Faruque, S.M. et al. Transmissibility of cholera: in vivo–formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. USA 103, 6350–6355 (2006).
Tamayo, R., Patimalla, B. & Camilli, A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 78, 3560–3569 (2010).
Schultz, J., Milpetz, F., Bork, P. & Ponting, C.P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95, 5857–5864 (1998).
Yeats, C., Bentley, S. & Bateman, A. New knowledge from old: in silico discovery of novel protein domains in Streptomyces coelicolor. BMC Microbiol. 3, 3 (2003).
Gadwal, S., Korotkov, K.V., Delarosa, J.R., Hol, W.G. & Sandkvist, M. Functional and structural characterization of Vibrio cholerae extracellular serine protease B, VesB. J. Biol. Chem. 289, 8288–8298 (2014).
Sikora, A.E., Zielke, R.A., Lawrence, D.A., Andrews, P.C. & Sandkvist, M. Proteomic analysis of the Vibrio cholerae type II secretome reveals new proteins, including three related serine proteases. J. Biol. Chem. 286, 16555–16566 (2011).
Siezen, R.J. & Leunissen, J.A. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6, 501–523 (1997).
Altindis, E., Fu, Y. & Mekalanos, J.J. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. USA 111, E1548–E1556 (2014).
Smith, D.R. et al. In situ proteolysis of the Vibrio cholerae matrix protein RbmA promotes biofilm recruitment. Proc. Natl. Acad. Sci. USA 112, 10491–10496 (2015).
Park, B.R. et al. A metalloprotease secreted by the type II secretion system links Vibrio cholerae with collagen. J. Bacteriol. 197, 1051–1064 (2015).
Chalfoun, N.R. et al. Purification and characterization of AsES protein: a subtilisin secreted by Acremonium strictum is a novel plant defense elicitor. J. Biol. Chem. 288, 14098–14113 (2013).
Mandlik, A. et al. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10, 165–174 (2011).
Fiore, A.E., Michalski, J.M., Russell, R.G., Sears, C.L. & Kaper, J.B. Cloning, characterization, and chromosomal mapping of a phospholipase (lecithinase) produced by Vibrio cholerae. Infect. Immun. 65, 3112–3117 (1997).
Pride, A.C., Herrera, C.M., Guan, Z., Giles, D.K. & Trent, M.S. The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. MBio 4, e00305–13 (2013).
Lundwall, A. & Brattsand, M. Kallikrein-related peptidases. Cell. Mol. Life Sci. 65, 2019–2038 (2008).
Holmes, R.S. & Cox, L.A. Comparative structures and evolution of vertebrate carboxyl ester lipase (CEL) genes and proteins with a major role in reverse cholesterol transport. Cholesterol 2011, 781643 (2011).
Pedersen, L.L. & Turco, S.J. Galactofuranose metabolism: a potential target for antimicrobial chemotherapy. Cell. Mol. Life Sci. 60, 259–266 (2003).
Wrackmeyer, U., Hansen, G.H., Seya, T. & Danielsen, E.M. Intelectin: a novel lipid raft-associated protein in the enterocyte brush border. Biochemistry 45, 9188–9197 (2006).
Pemberton, A.D. et al. Innate BALB/c enteric epithelial responses to Trichinella spiralis: inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J. Immunol. 173, 1894–1901 (2004).
Voehringer, D. et al. Nippostrongylus brasiliensis: identification of intelectin-1 and -2 as Stat6-dependent genes expressed in lung and intestine during infection. Exp. Parasitol. 116, 458–466 (2007).
Ellis, C.N. et al. Comparative proteomic analysis reveals activation of mucosal innate immune signaling pathways during cholera. Infect. Immun. 83, 1089–1103 (2015).
Tsuji, S. et al. Capture of heat-killed Mycobacterium bovis bacillus Calmette-Guérin by intelectin-1 deposited on cell surfaces. Glycobiology 19, 518–526 (2009).
Wesener, D.A. et al. Recognition of microbial glycans by human intelectin-1. Nat. Struct. Mol. Biol. 22, 603–610 (2015).
Varina, M., Denkin, S.M., Staroscik, A.M. & Nelson, D.R. Identification and characterization of Epp, the secreted processing protease for the Vibrio anguillarum EmpA metalloprotease. J. Bacteriol. 190, 6589–6597 (2008).
Bracco, U. Effect of triglyceride structure on fat absorption. Am. J. Clin. Nutr. 60 (suppl. 6): 1002S–1009S (1994).
Drzazga, A., Sowińska, A. & Koziołkiewicz, M. Lysophosphatidylcholine and lysophosphatidylinosiol—novel promising signaling molecules and their possible therapeutic activity. Acta Pol. Pharm. 71, 887–899 (2014).
Suzuki, Y.A., Shin, K. & Lönnerdal, B. Molecular cloning and functional expression of a human intestinal lactoferrin receptor. Biochemistry 40, 15771–15779 (2001).
Yang, R.Z. et al. Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action. Am. J. Physiol. Endocrinol. Metab. 290, E1253–E1261 (2006).
Hatzios, S.K., Ringgaard, S., Davis, B.M. & Waldor, M.K. Studies of dynamic protein-protein interactions in bacteria using Renilla luciferase complementation are undermined by nonspecific enzyme inhibition. PLoS ONE 7, e43175 (2012).
Abel, S. & Waldor, M.K. Infant rabbit model for diarrheal diseases. Curr. Protoc. Microbiol. 38, 6A.6.1–6A.6.15 (2015).
Vizcaíno, J.A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014).
Kamat, S.S. et al. Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay. Nat. Chem. Biol. 11, 164–171 (2015).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Breslow, N.E. & Clayton, D.G. Approximate interference in generalized linear mixed models. J. Am. Stat. Assoc. 88, 9–25 (1993).
Wolfinger, R. & O'Connell, M. Generalized linear mixed models: a pseudo-likelihood approach. J. Stat. Comput. Simul. 48, 233–243 (1993).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Venables, W.N. & Ripley, B.D. Modern Applied Statistics with S 4th edn. (Springer, 2002).
Acknowledgements
We gratefully acknowledge R. LaRocque, Y. Millet and J. Lee for reagents and technical assistance, and members of M.K.W.'s lab for helpful discussions. We also thank V. Carey for the statistical analysis of MS data and D. Bak for help formatting MS data sets. This work was supported by the National Institutes of Health (R37 AI-042347 to M.K.W., F31 AI-120665 to T.H., R01 AI-106878 and U01 AI-058935 to E.T.R., Institutional Training Grant T32 DK 7477-30 to S.K.H.), the Howard Hughes Medical Institute (M.K.W.), the Charles A. King Trust Postdoctoral Fellowship Program, Bank of America, N.A., Co-Trustee (S.K.H.), the Damon Runyon Cancer Research Foundation (DRR-18-12 to E.W.), the Smith Family Foundation (E.W.) and the Swiss National Science Foundation (P300P3_155287/1 to S.A.).
Author information
Authors and Affiliations
Contributions
S.K.H., E.W. and M.K.W. conceived the project and designed the experiments. S.K.H. generated the V. cholerae mutant strains, performed all biochemistry and ABPP experiments and analyzed the MS data. S.A., T.H. and D.M. performed the rabbit infections. J.M. performed the MS experiments and assisted with MS data analysis. J.S. provided technical guidance on intelectin assays. L.C. assisted with the construction of V. cholerae mutant strains. D.A.B. synthesized FP-biotin. F.Q. and E.T.R. provided the human choleric stool used in this study. S.K.H., B.M.D., and M.K.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–9 and Supplementary Figures 1–10. (PDF 7161 kb)
Supplementary Data Set 1
FP-biotin-enriched proteins detected in wild-type cecal fluid, comparative ABPP-MudPIT analysis of wild-type and S361A cecal fluid and combined unfiltered data from all ABPP-MudPIT analyses of wild-type cecal fluid. (XLSX 92 kb)
Supplementary Data Set 2
FP-biotin–enriched proteins from human choleric stool supernatant and unfiltered data from ABPP-MudPIT analysis of human choleric stool supernatant. (XLSX 28 kb)
Supplementary Data Set 3
ABPP-MudPIT analysis of wild-type and ΔivaP biofilm culture supernatants. (XLSX 23 kb)
Supplementary Data Set 4
ABPP-MudPIT analysis of exponential- and stationary-phase V. cholerae cell lysates. (XLSX 40 kb)
Supplementary Data Set 5
Comparative total and free fatty acid analysis of wild-type and Δquad cecal fluid. (XLSX 21 kb)
Supplementary Data Set 6
V. cholerae and rabbit proteins detected in unfractionated wild-type and Δquad cecal fluid. (XLSX 188 kb)
Supplementary Data Set 7
V. cholerae and human proteins detected in unfractionated human choleric stool. (XLSX 52 kb)
Source data
Rights and permissions
About this article
Cite this article
Hatzios, S., Abel, S., Martell, J. et al. Chemoproteomic profiling of host and pathogen enzymes active in cholera. Nat Chem Biol 12, 268–274 (2016). https://doi.org/10.1038/nchembio.2025
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2025
This article is cited by
-
An infection-induced oxidation site regulates legumain processing and tumor growth
Nature Chemical Biology (2022)
-
Proteomic analysis of the host–pathogen interface in experimental cholera
Nature Chemical Biology (2021)
-
R-spondin-3 induces secretory, antimicrobial Lgr5+ cells in the stomach
Nature Cell Biology (2019)
-
Molecular insights into Vibrio cholerae’s intra-amoebal host-pathogen interactions
Nature Communications (2018)
-
Identification of a S. aureus virulence factor by activity-based protein profiling (ABPP)
Nature Chemical Biology (2018)